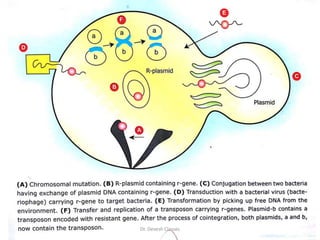
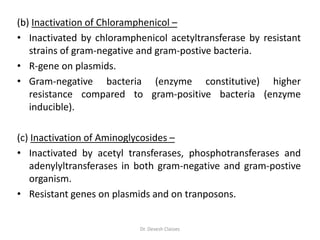
The document discusses bacterial drug resistance, detailing concepts such as intrinsic and acquired resistance, heteroresistance, and genetic mechanisms contributing to resistance. It outlines various methods of resistance transfer among bacteria, such as conjugation, transduction, and transformation, as well as biochemical mechanisms that lead to antibiotic inactivation. Finally, the conclusion highlights the role of evolution and clinical practices in the emergence of resistance, emphasizing the need for cautious antibiotic use and targeted treatment strategies.