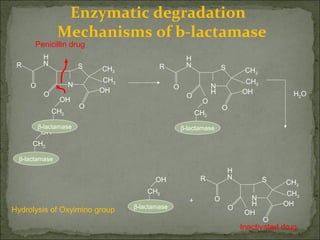
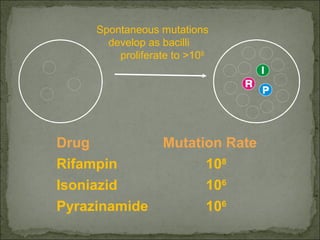
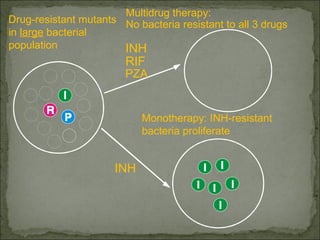
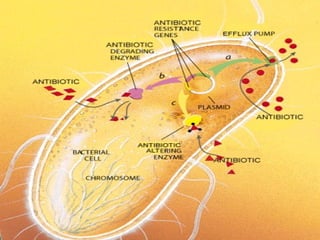
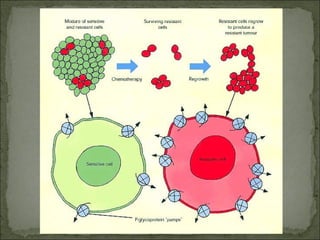
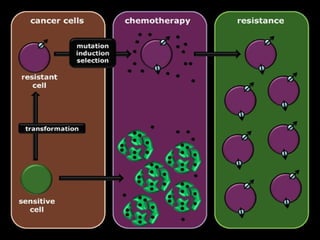
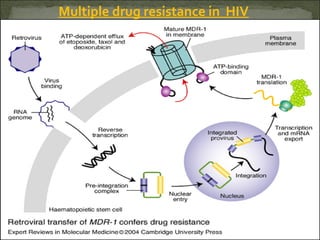
This document discusses multiple drug resistance (MDR) in several organisms and contexts. It begins with an introduction to MDR and its mechanisms, including enzymatic degradation, mutations, efflux pumps, and decreased membrane permeability. Specific examples of MDR are then explored in tuberculosis, bacteria, cancer cells, HIV, and malaria. The mechanisms of resistance and genes involved vary by organism but often involve efflux pump proteins like P-glycoprotein. MDR is an increasing public health issue due to its role in antibiotic resistance.