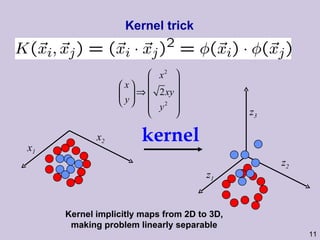
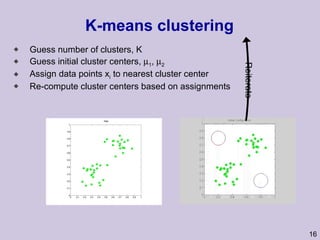
This document provides an overview and introduction to machine learning and probabilistic graphical models. It discusses key topics such as supervised learning, unsupervised learning, graphical models, inference, and structure learning. The document covers techniques like decision trees, neural networks, clustering, dimensionality reduction, Bayesian networks, and learning the structure of probabilistic graphical models.















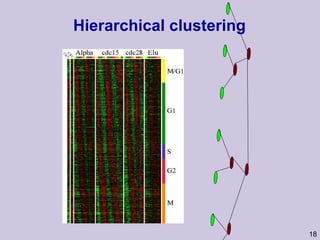
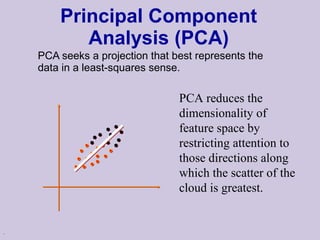
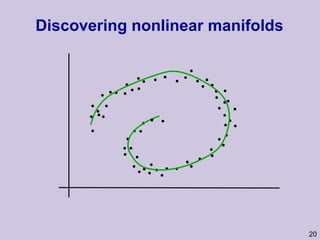
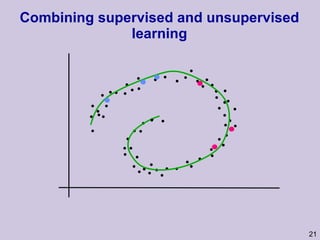
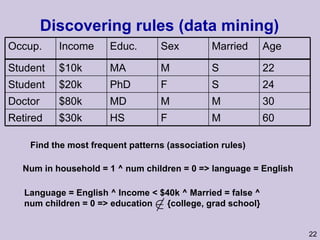

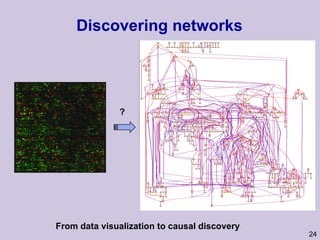



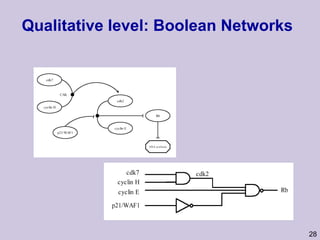



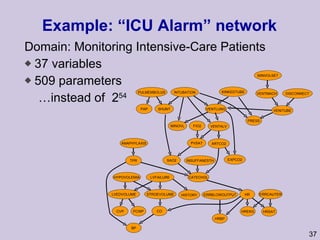
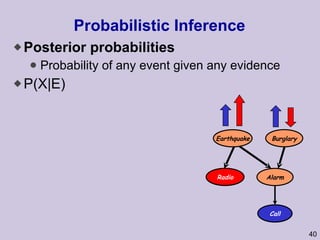

















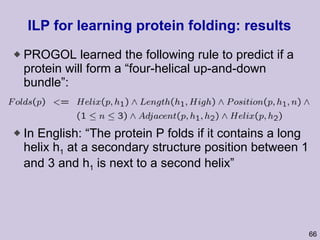









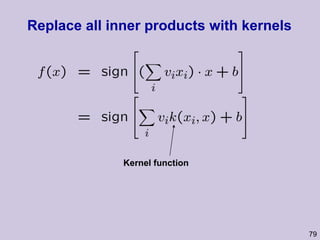




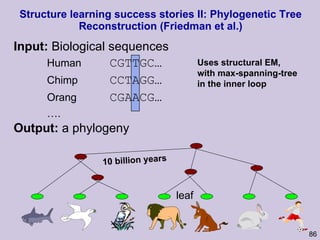
![Structure learning success stories: gene regulation network (Friedman et al.) Yeast data [Hughes et al 2000] 600 genes 300 experiments](https://image.slidesharecdn.com/an-introduction-to-machine-learning-and-probabilistic2915/85/An-introduction-to-machine-learning-and-probabilistic-85-320.jpg)