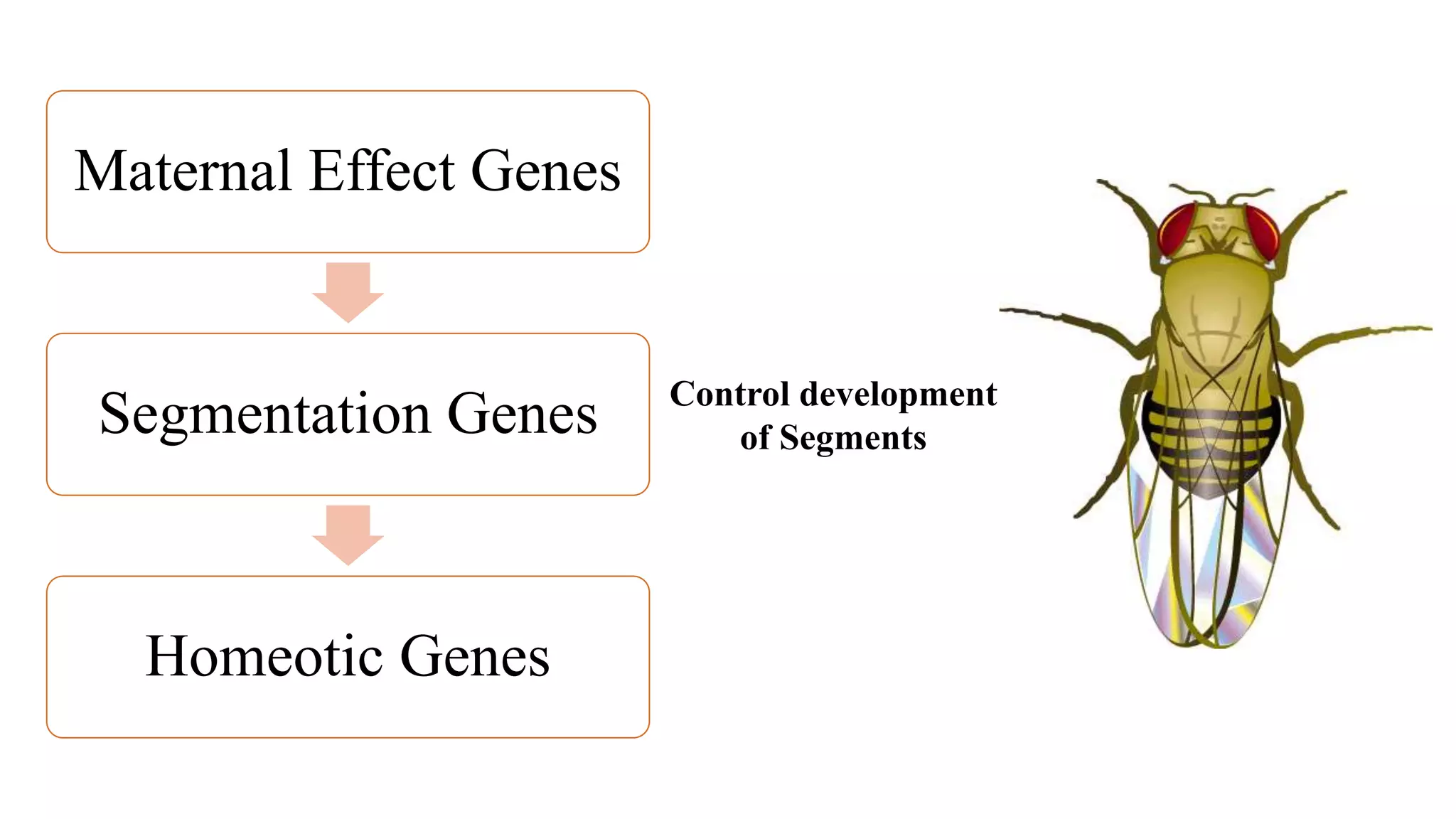
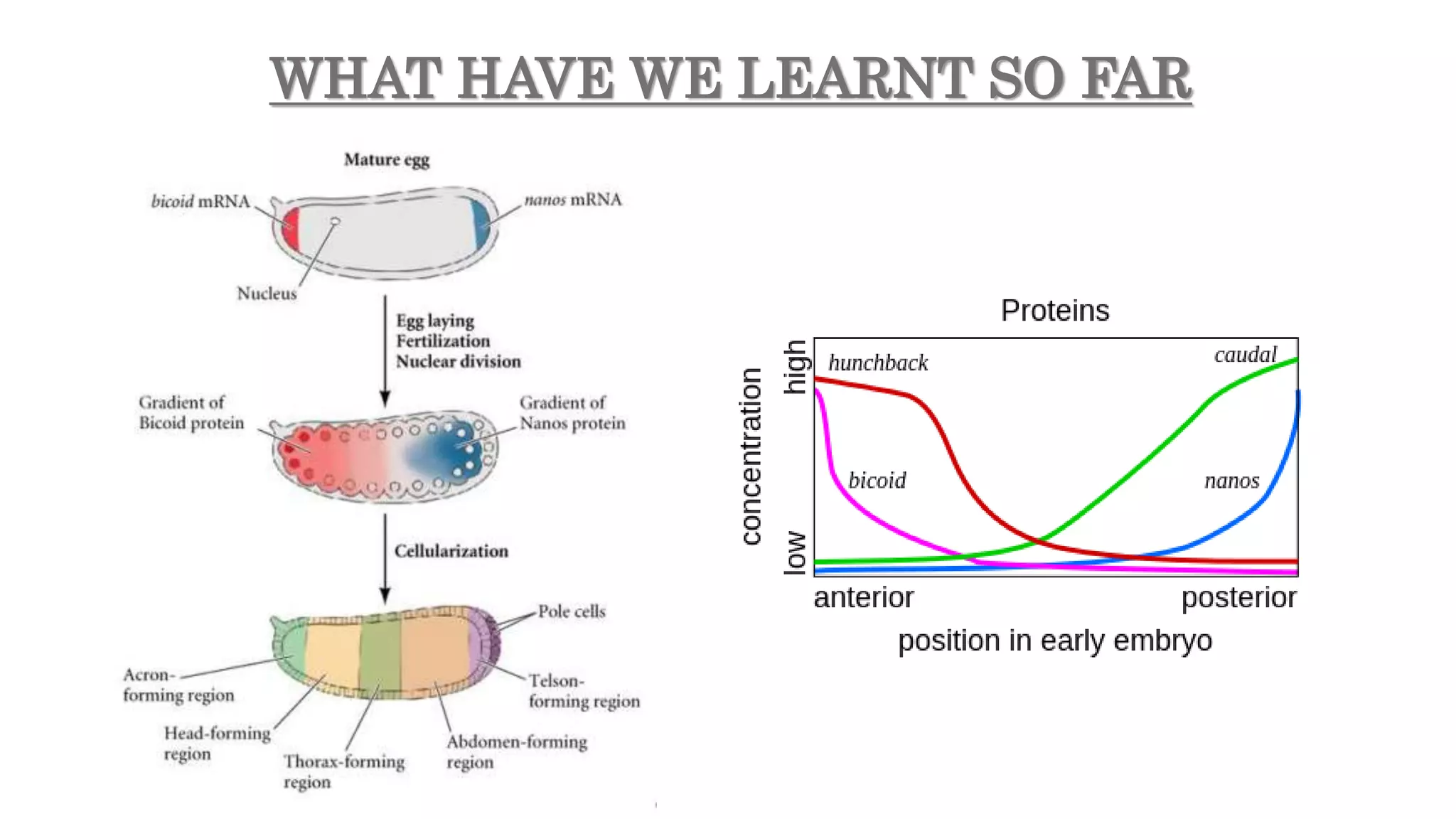
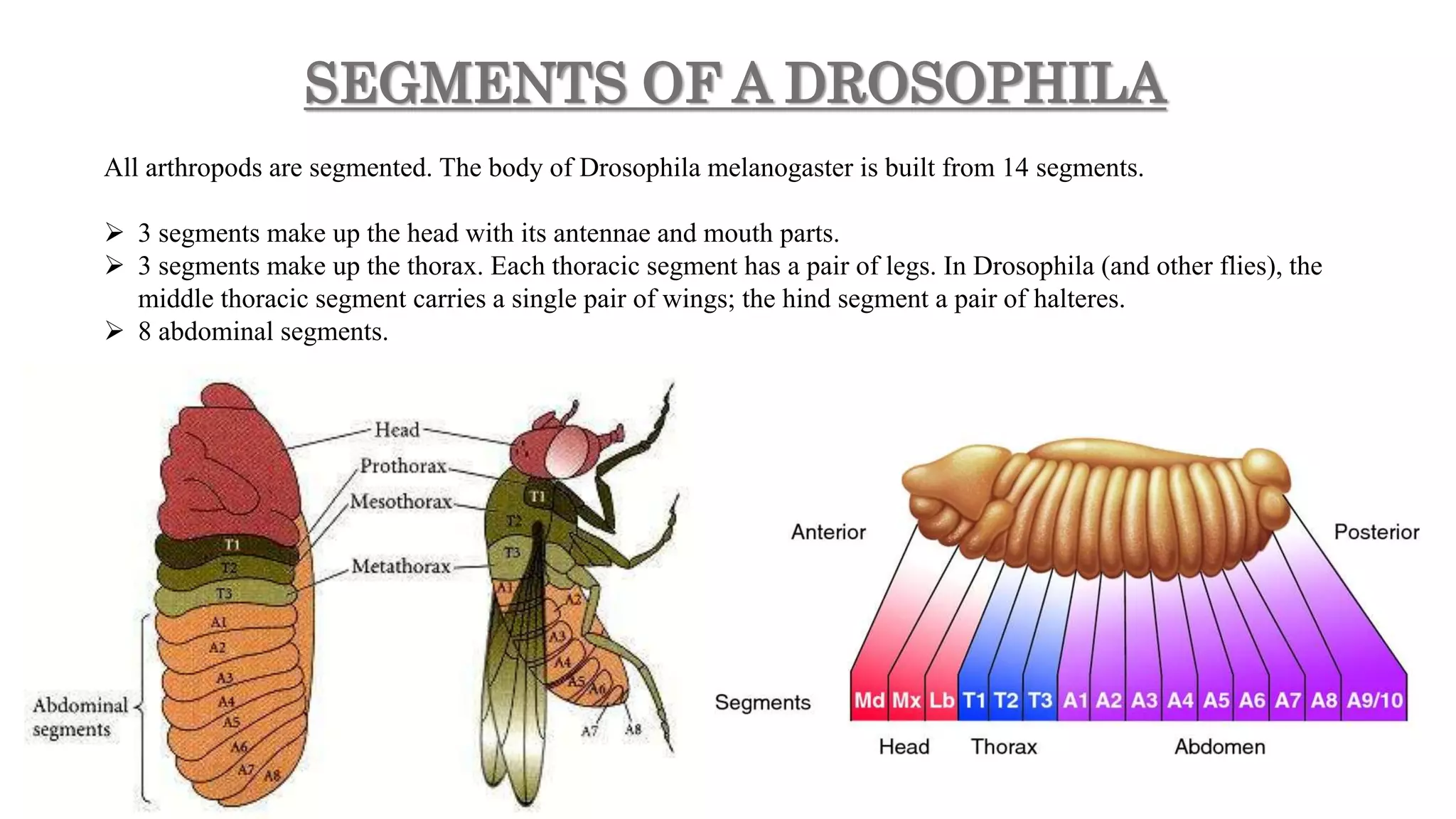
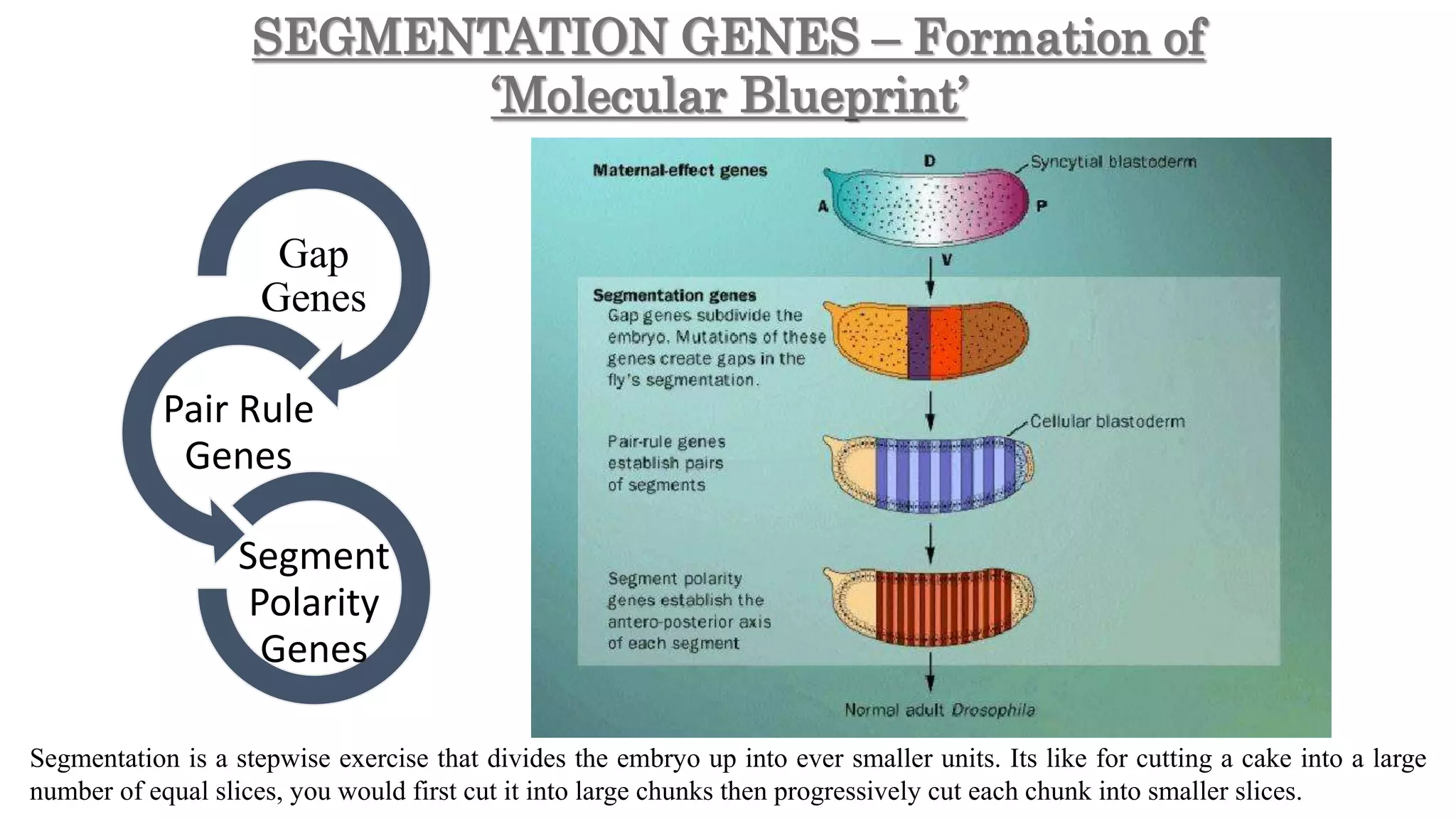
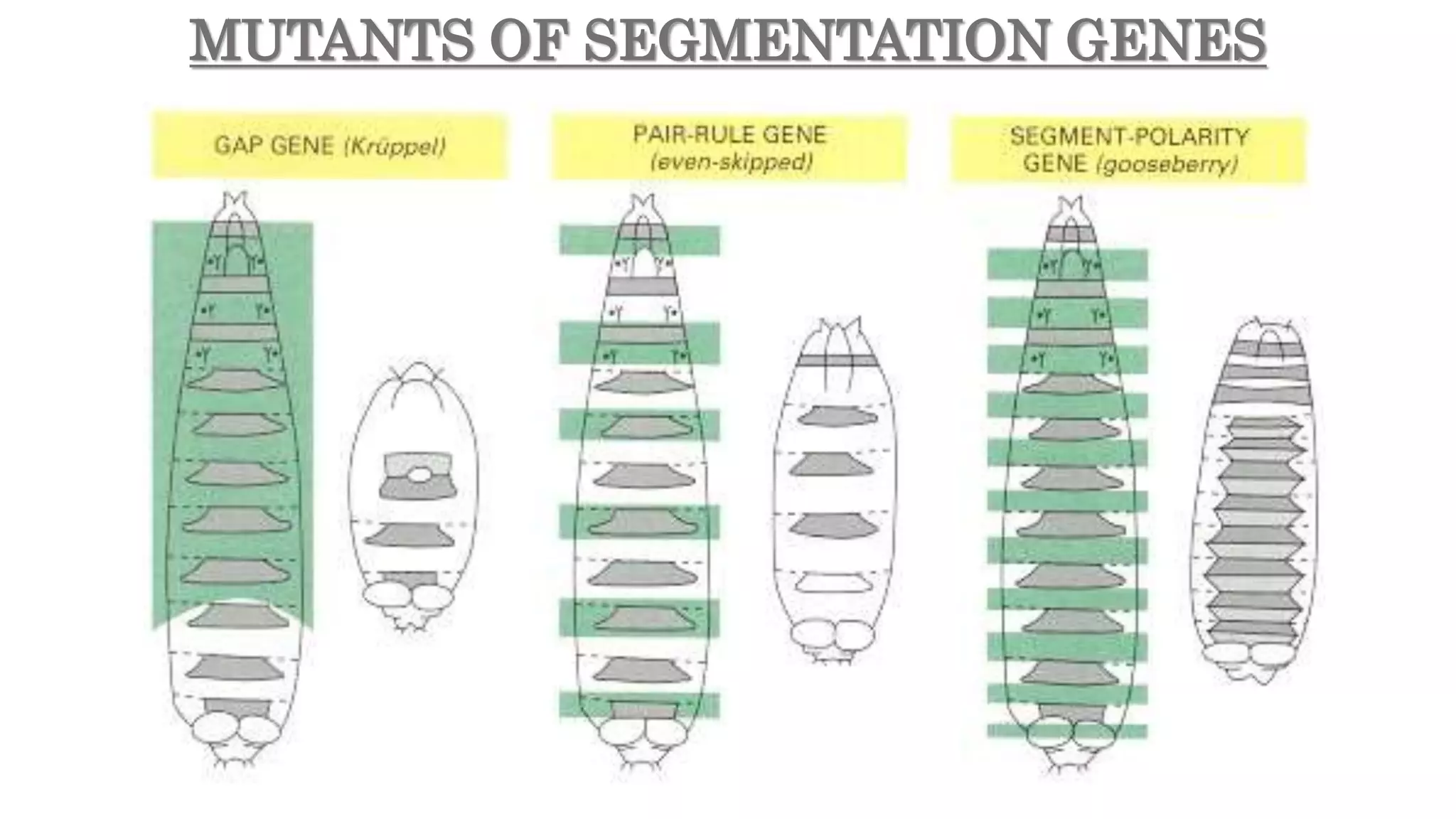
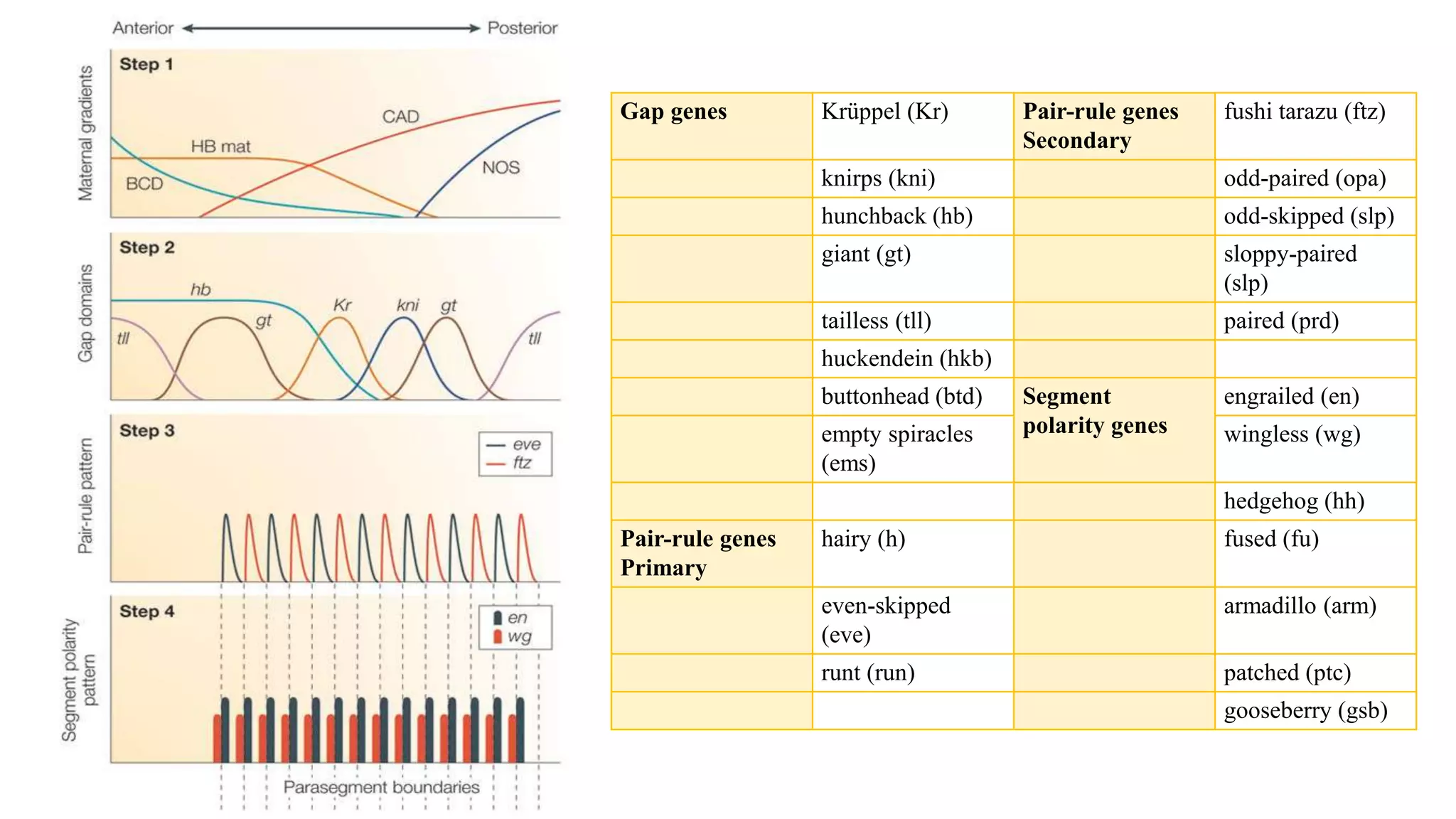
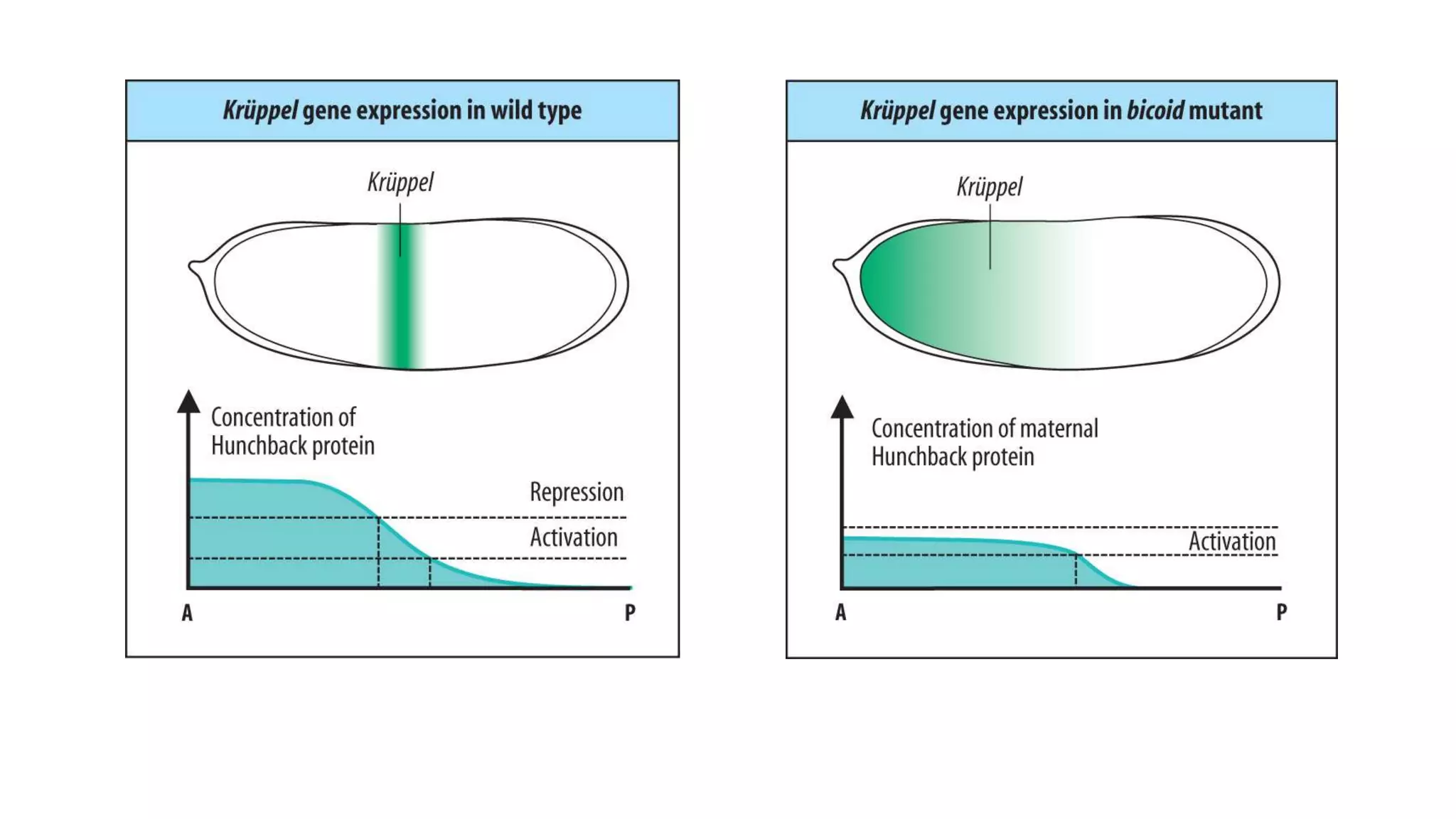
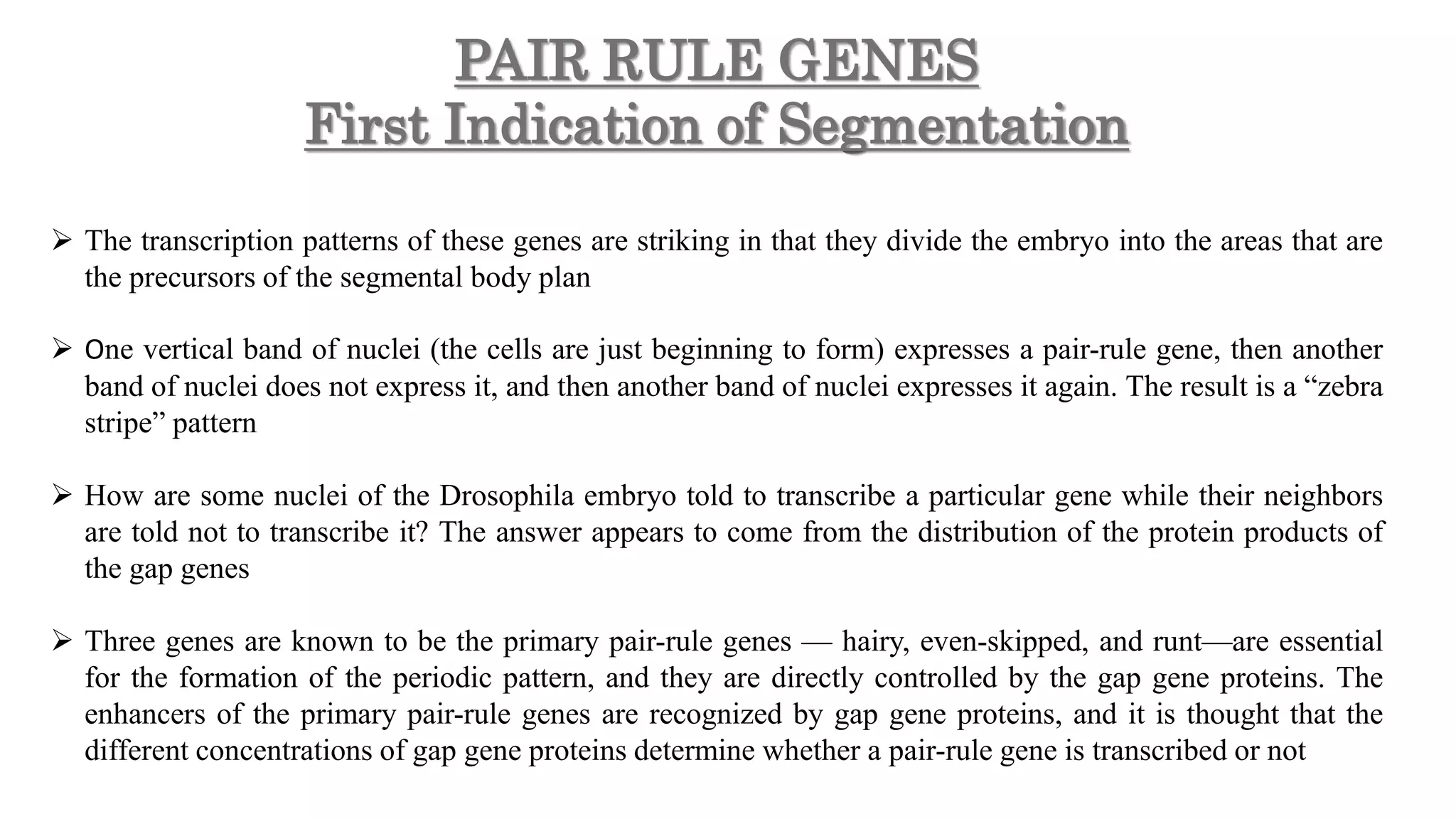
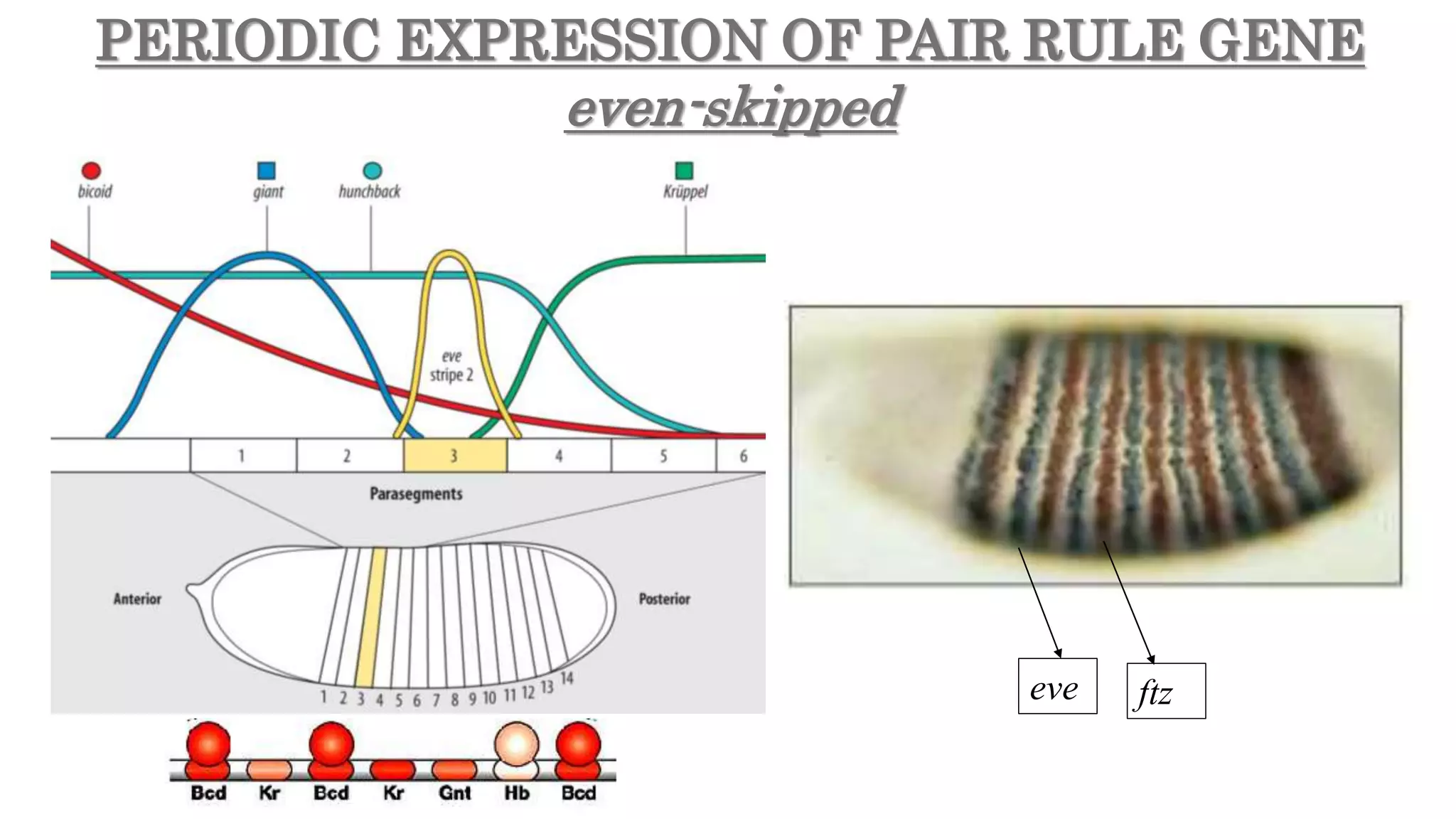
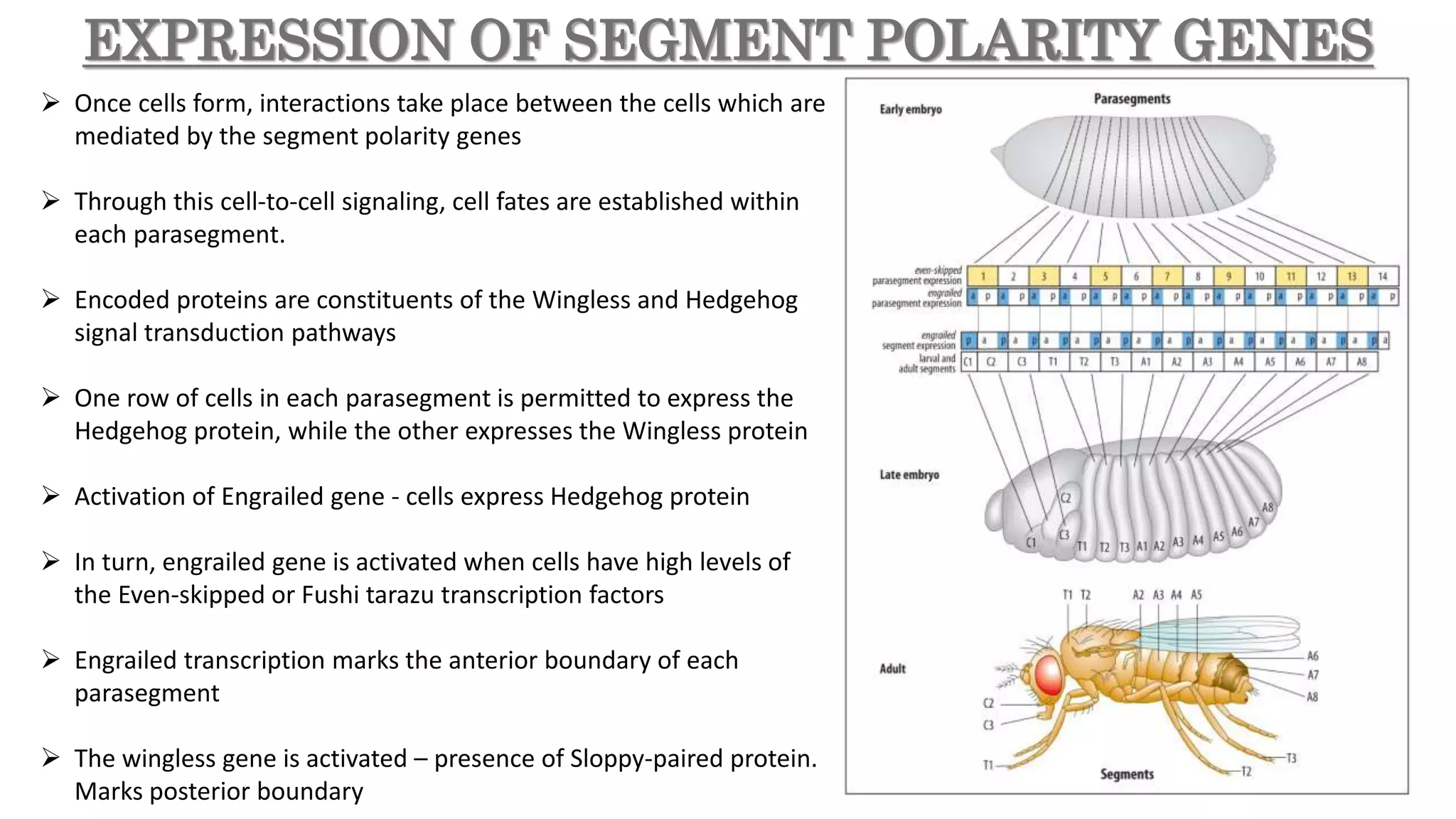
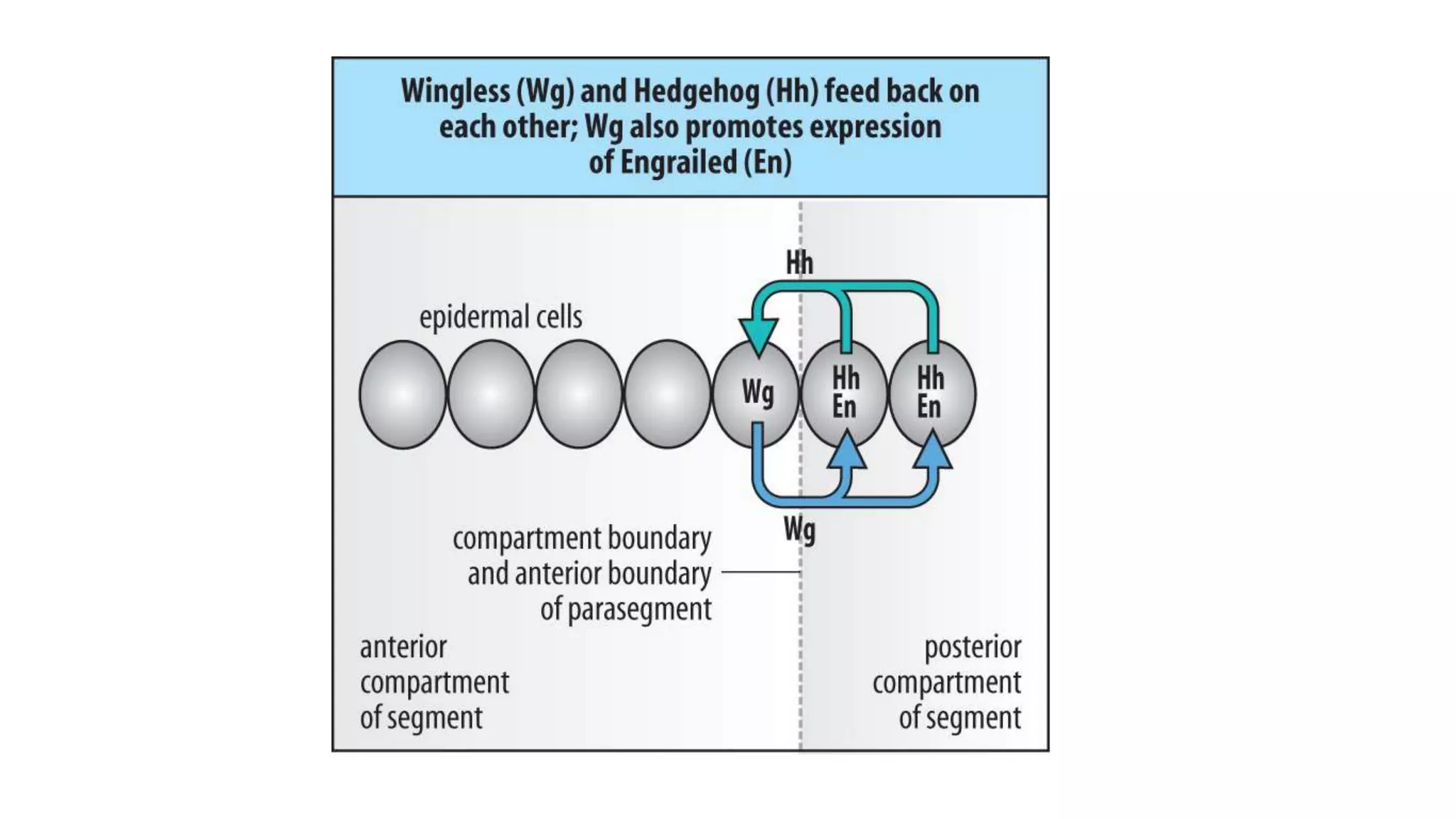
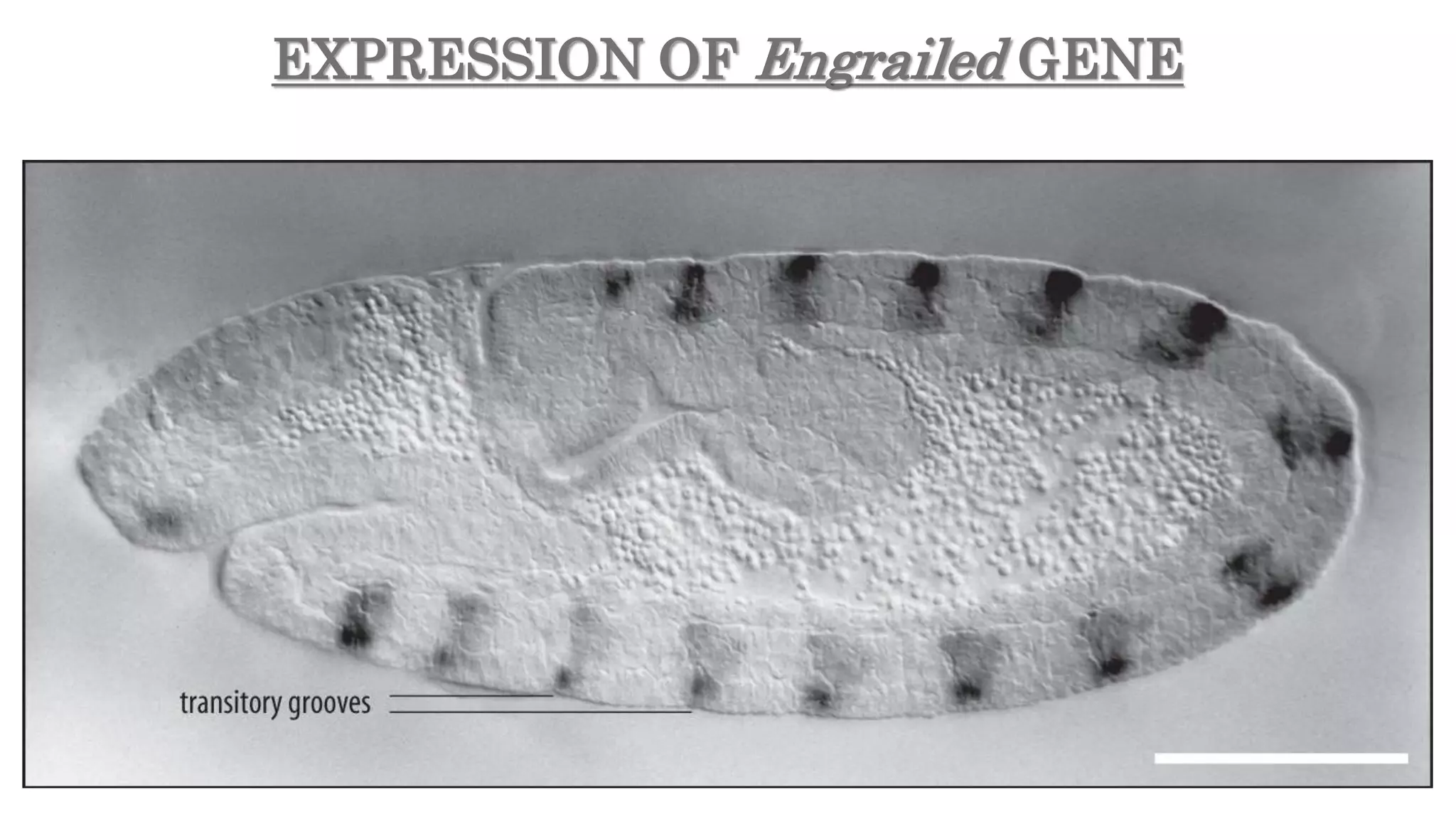
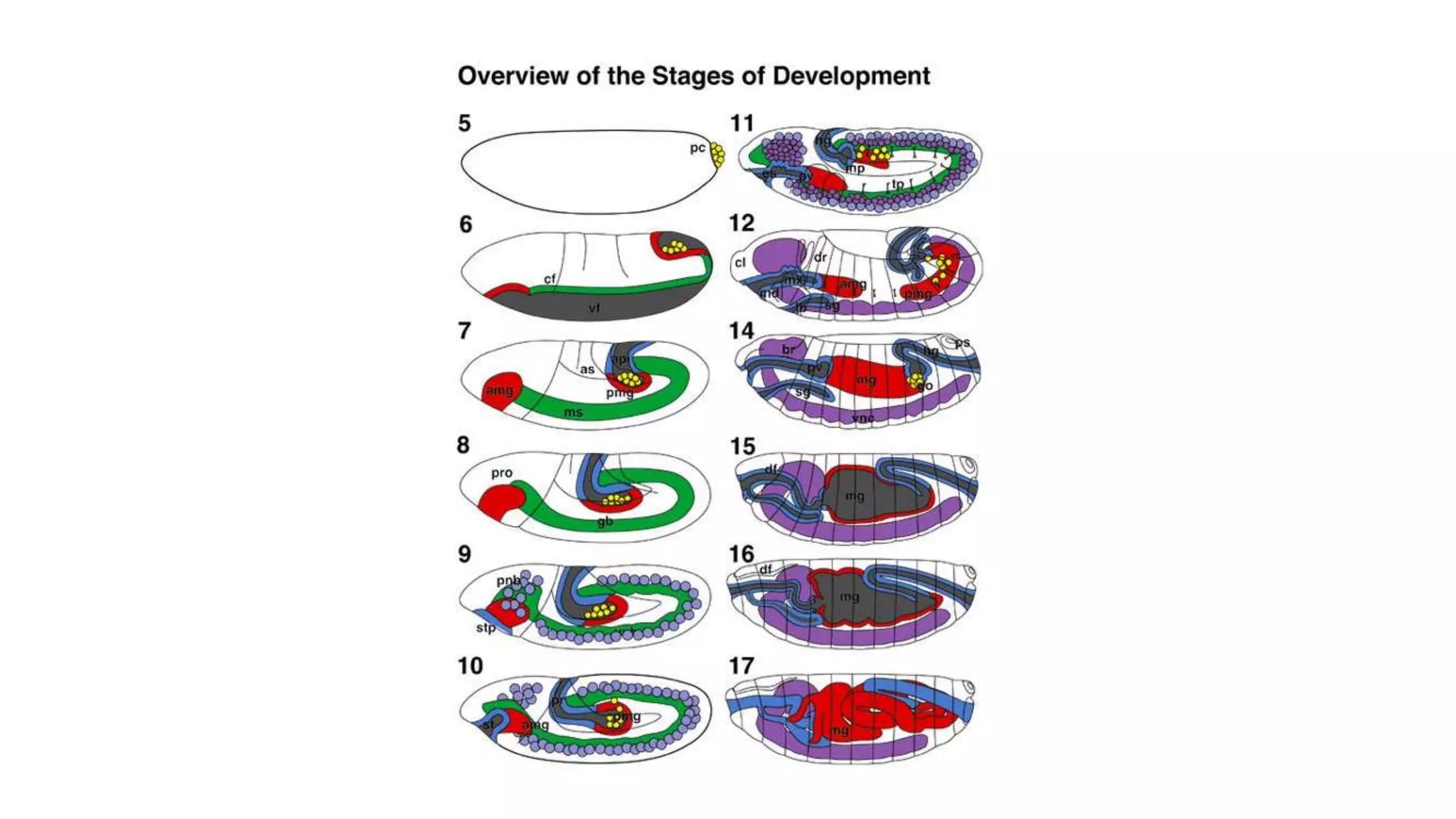
The document discusses the role of various genes in the development of Drosophila melanogaster, emphasizing segmentation and patterning mechanisms. It details how maternal effect, gap, pair-rule, and segment polarity genes interact to establish the segmented body plan through a stepwise process. The transcription patterns of these genes influence cell fate and the formation of structures such as legs and wings by using concentration gradients of proteins and signaling pathways.