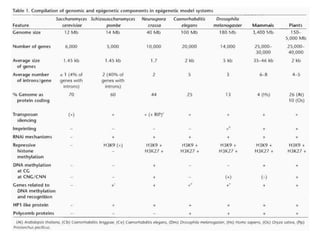
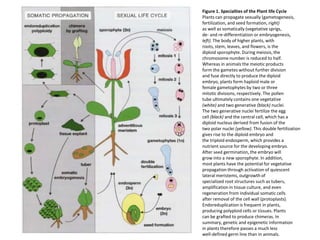
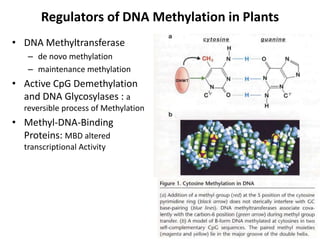
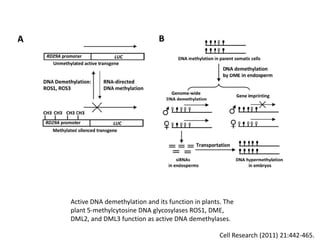
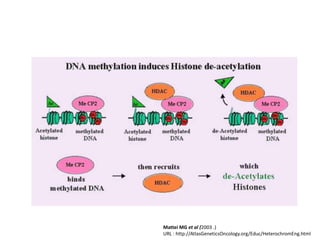
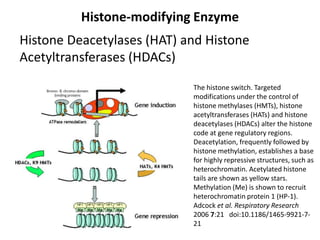
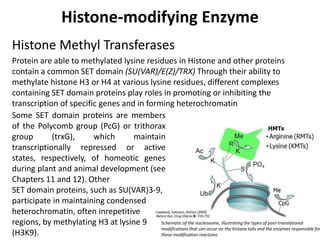
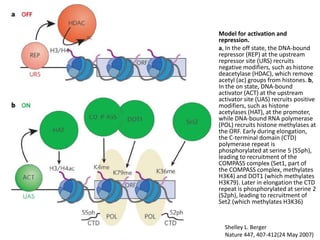
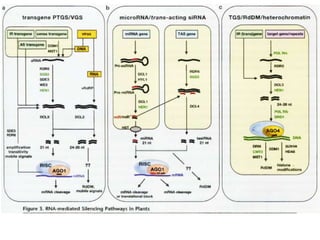
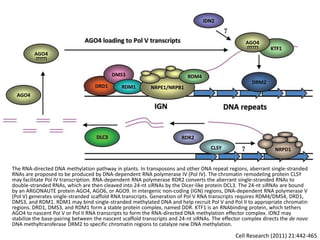
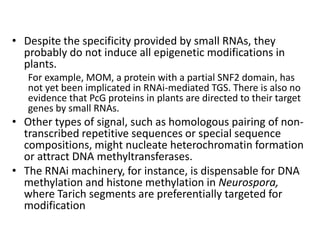
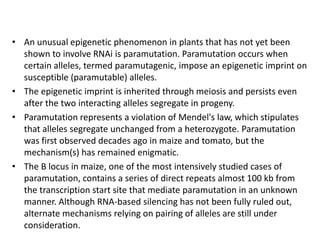
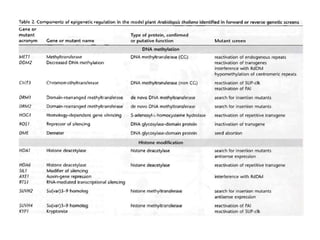
This document discusses epigenetic regulation in plants. It begins by outlining the benefits of studying epigenetics in plants, including their haploid stage and ability to tolerate polyploidy. It then describes the molecular components that regulate chromatin structure in plants, including DNA methyltransferases, histone-modifying enzymes, and other chromatin proteins. Next, it examines the RNA interference pathways involved in epigenetic gene silencing in plants. The document also notes that some epigenetic regulation occurs without RNA involvement, such as paramutation. Finally, it discusses how studying plant epigenetics can provide insights into human evolution and epigenetic reprogramming.