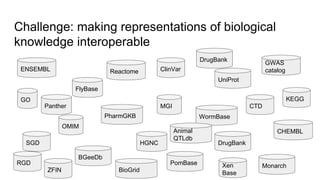
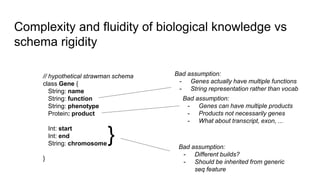
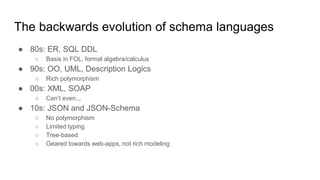
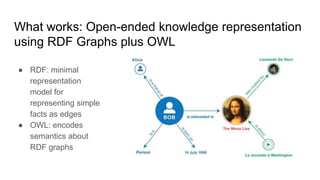
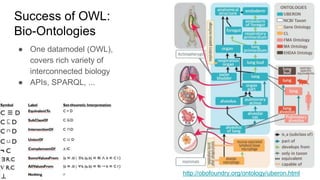
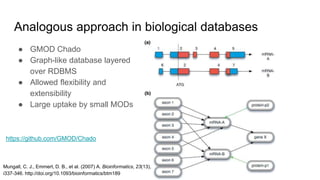
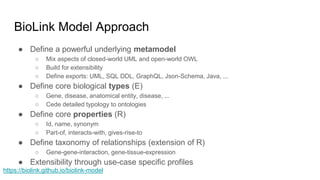
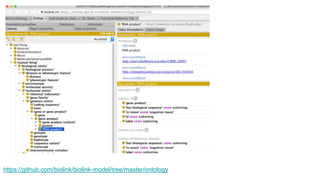
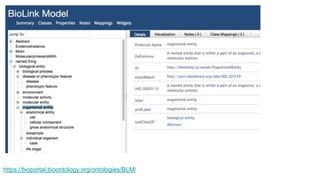
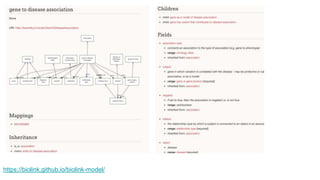
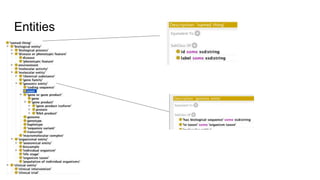
The document discusses the Biolink Model as a standardized schema for representing biological knowledge, addressing challenges of interoperability among various databases. It emphasizes the distinction between data and knowledge, advocates for extensibility in biological representation through RDF and OWL, and outlines the Biolink Model's approach to defining core biological types and relationships. Additionally, it notes the importance of flexibility in knowledge representation while maintaining necessary constraints for software systems.