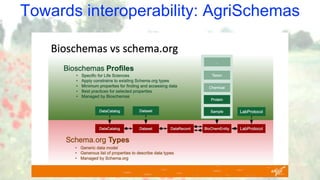
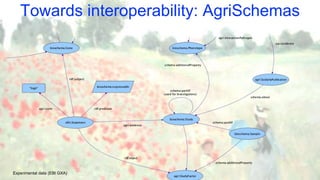
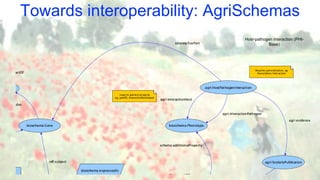
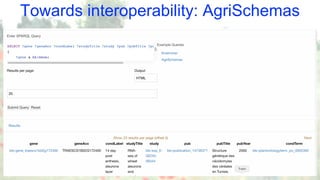
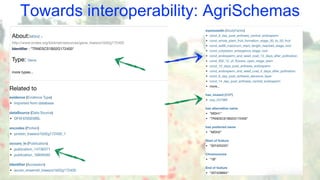
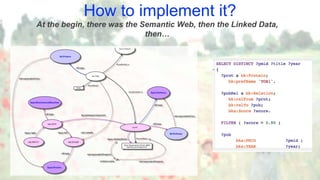
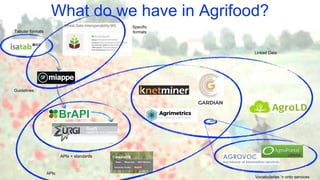
The document discusses the importance of sharing machine-readable data in the agrifood and life sciences sectors, emphasizing the need for interoperability through common formats and standards. It highlights examples like Knetminer and RDF/OWL as pivotal tools for data representation while pointing out challenges related to integration and the evolution of data querying approaches. The document concludes with suggestions for further development and exploration of existing schemas and endpoints to enhance data accessibility and usefulness.







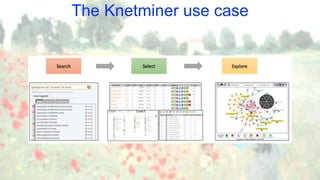
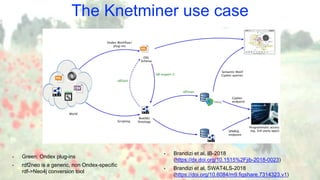
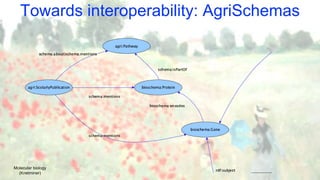
![The Knetminer use case
Cypher examples:
MATCH
// branching via ‘|’
(prot:Protein) - [:produced_by|consumed_by] -> (:Reaction)
// variable-length chains
- [:part_of*1..3] -> (pway:Path)
RETURN
prot.name, pwy LIMIT 1000
// Very compact forms available:
MATCH (prot:Protein) - (pway:Path) RETURN pway
• RDF + OWL used as a standardised modelling/representation language
(see BioKNO ontology: github.com/Rothamsted/bioknet-onto)
• SPARQL available too, both having pros/cons
(see our benchmark: github.com/Rothamsted/graphdb-benchmarks)
• Cypher being used for “Semantic Motif” queries, linking genes to entities of interest
(work in progress)](https://image.slidesharecdn.com/ib-2019-brandizi-agridata-where-we-are-190917223149/85/AgriFood-Data-Models-Standards-Tools-Use-Cases-15-320.jpg)