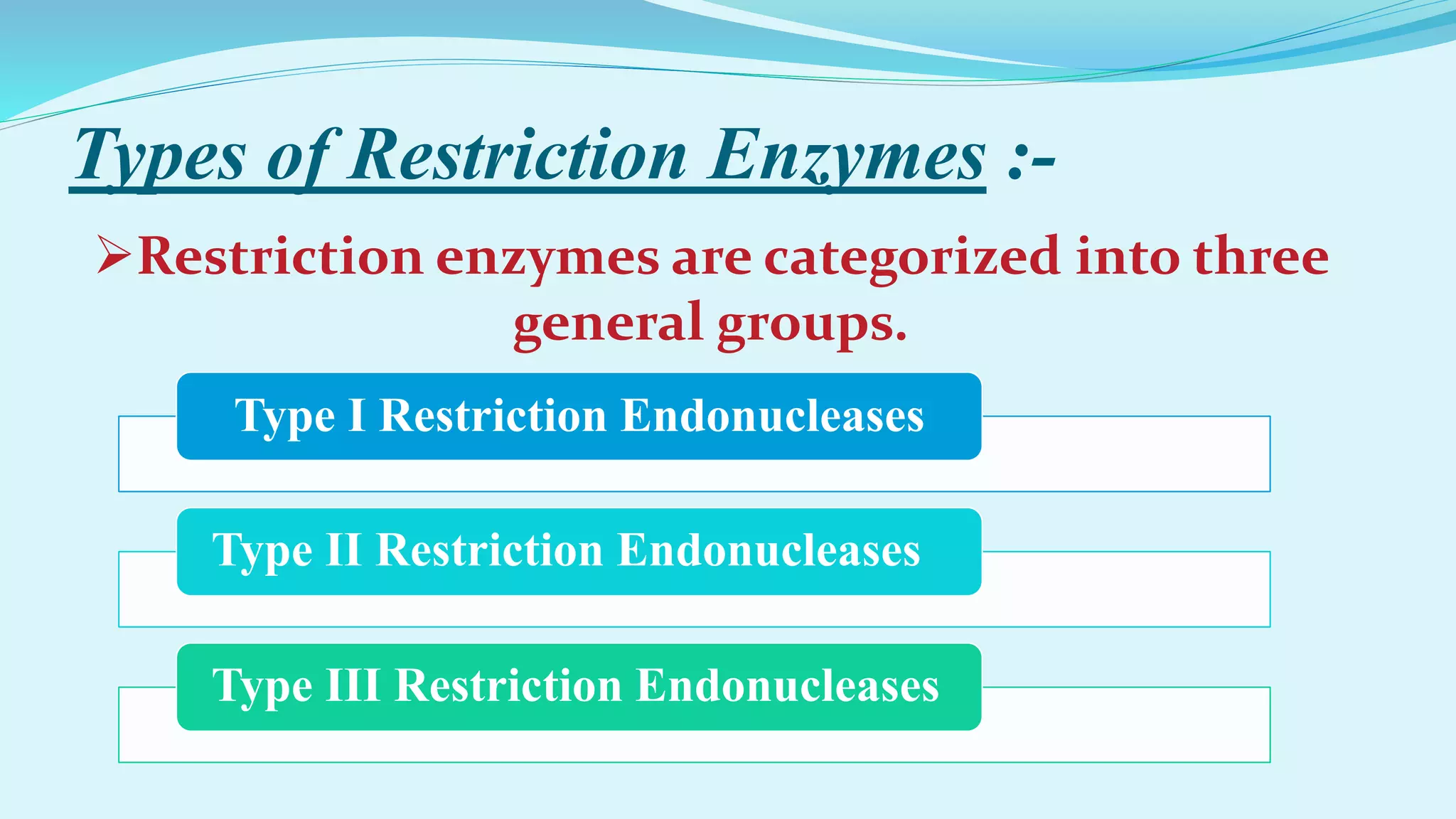
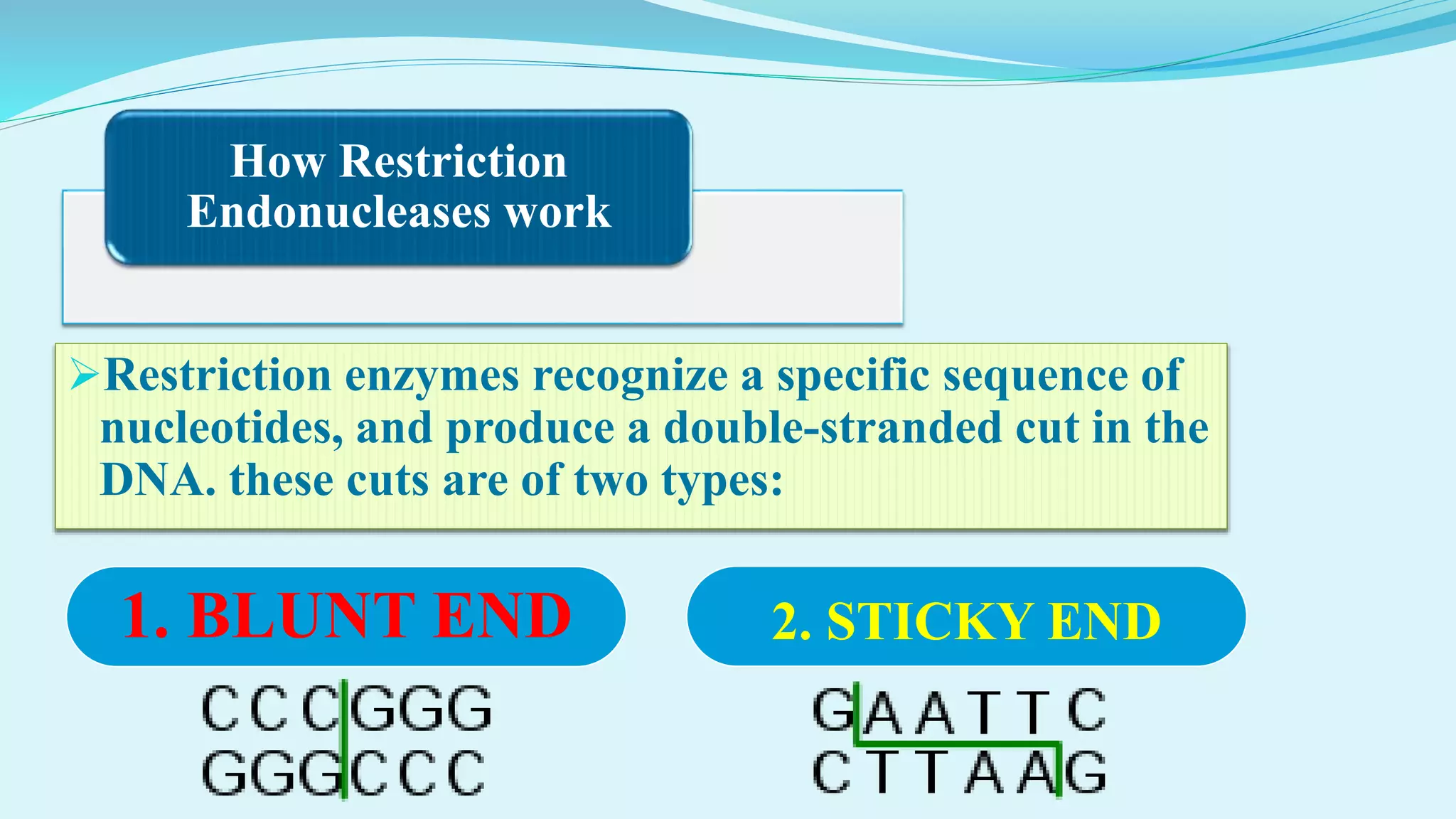
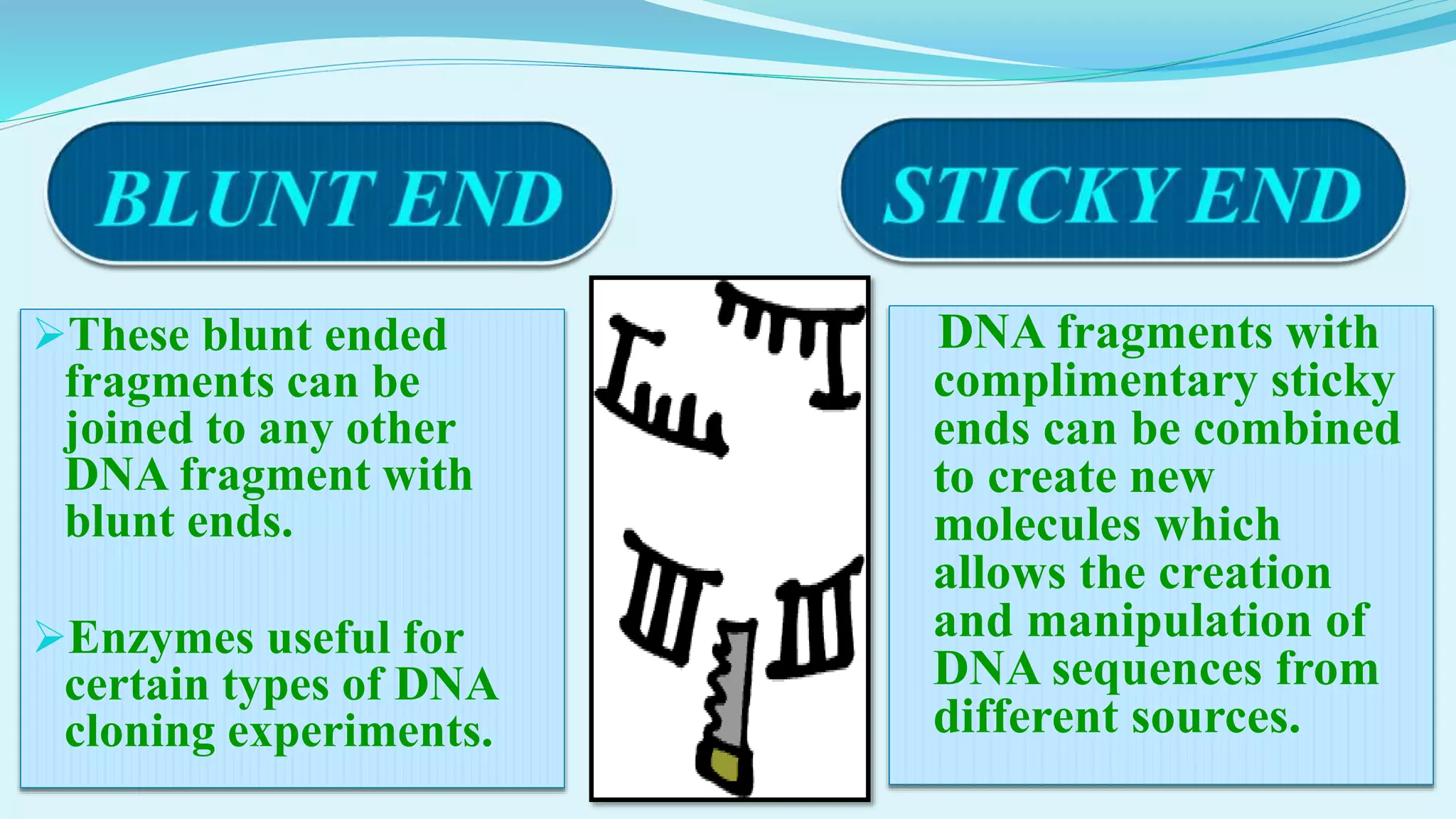
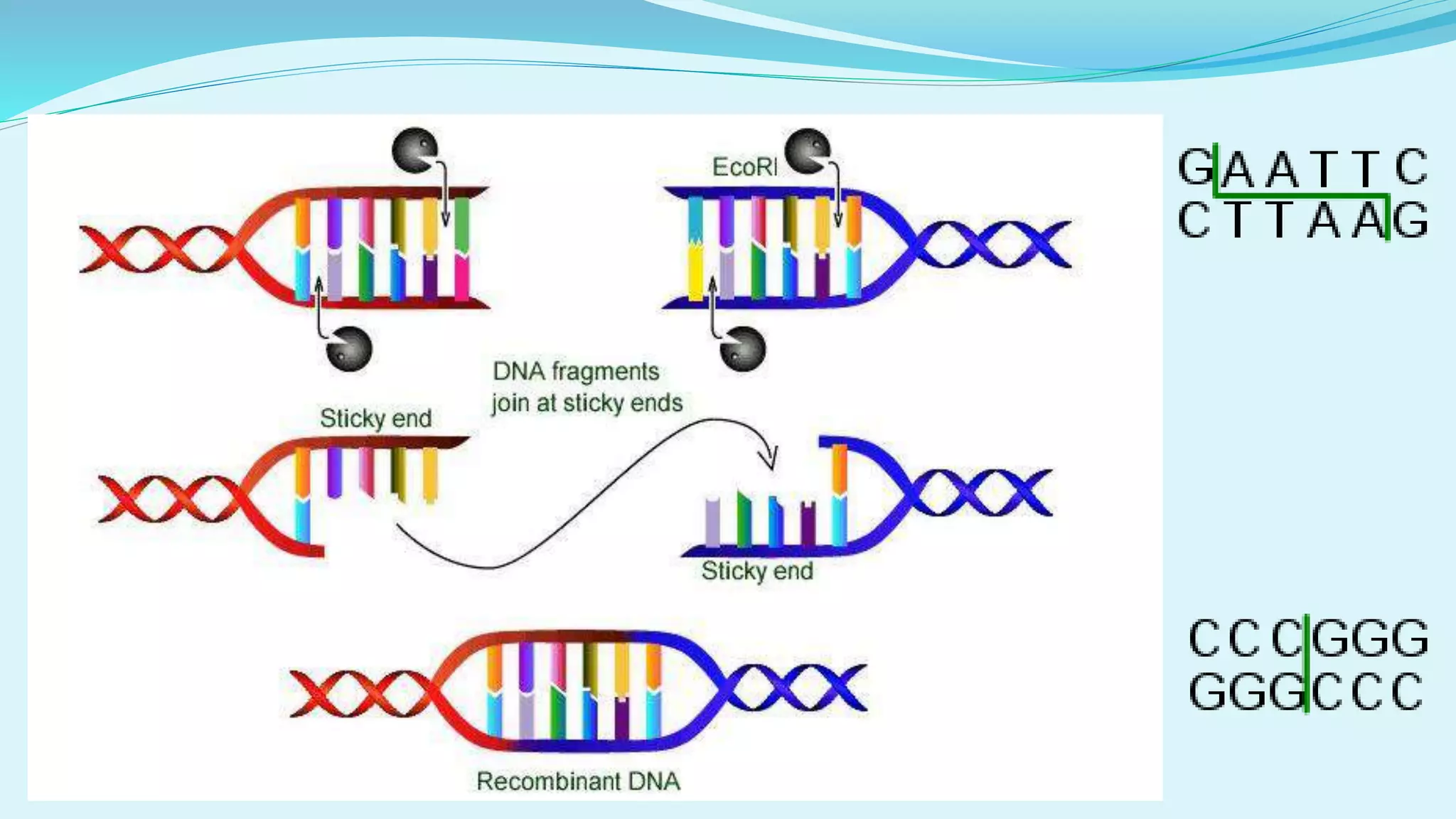
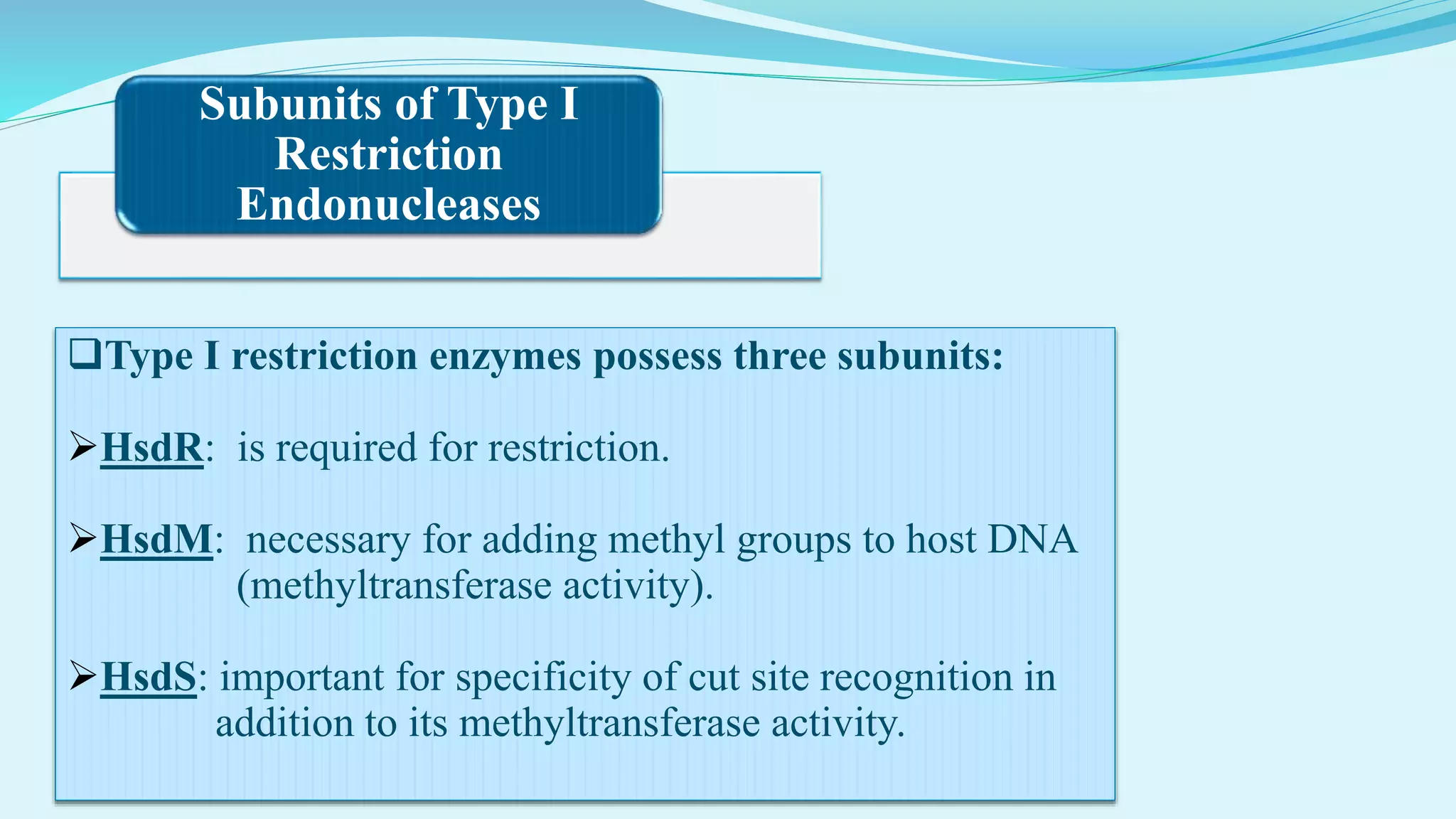
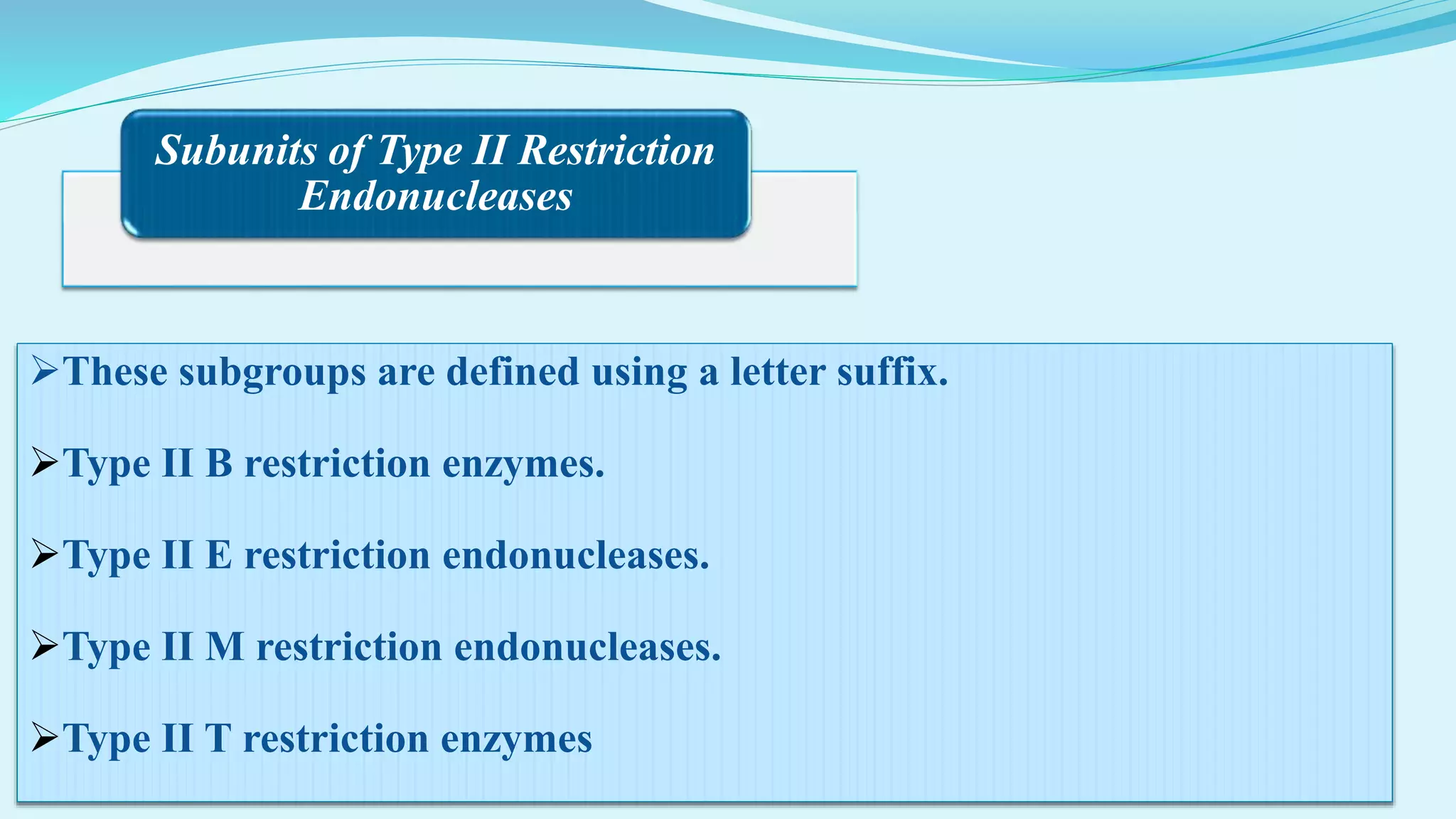
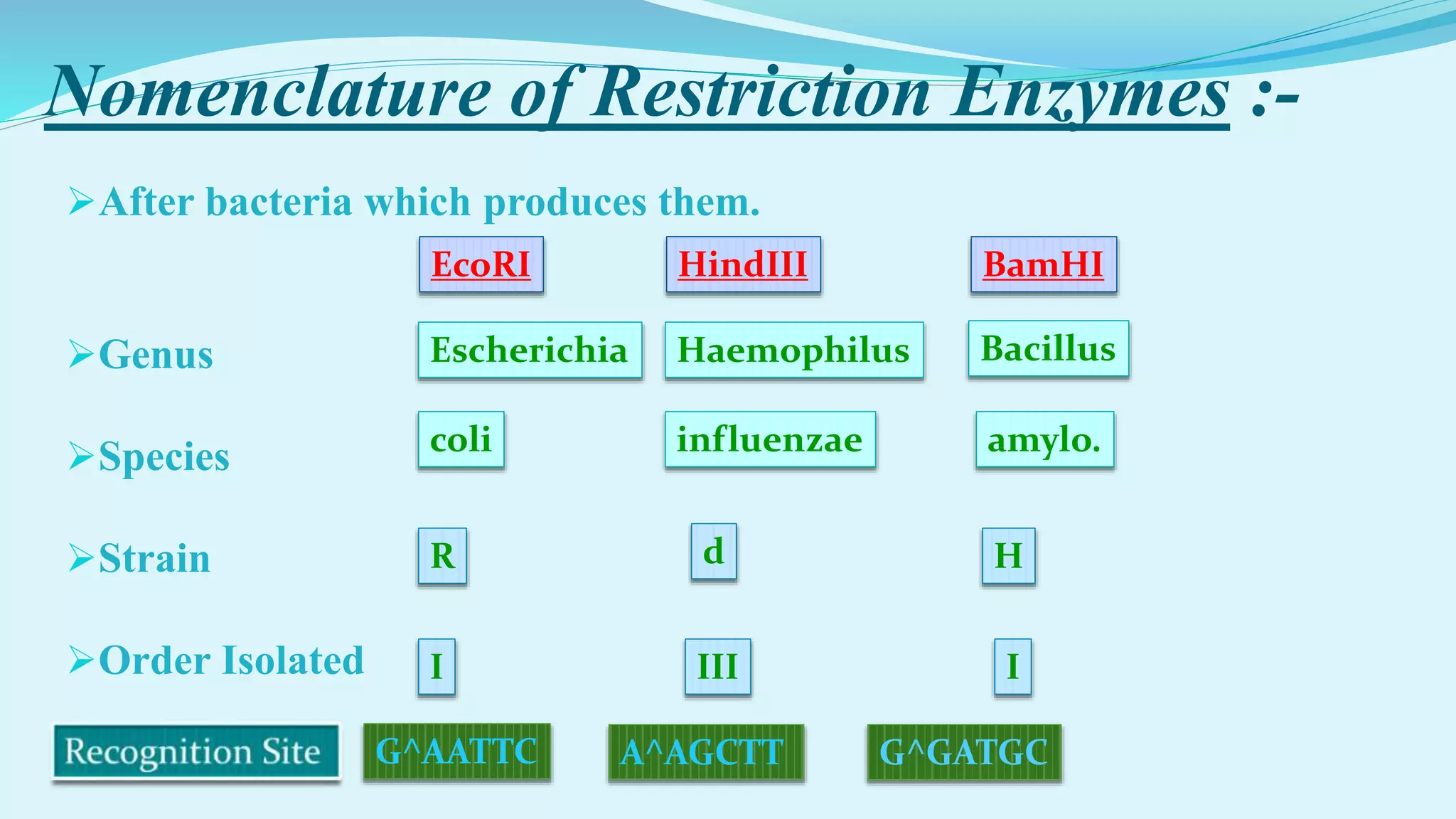
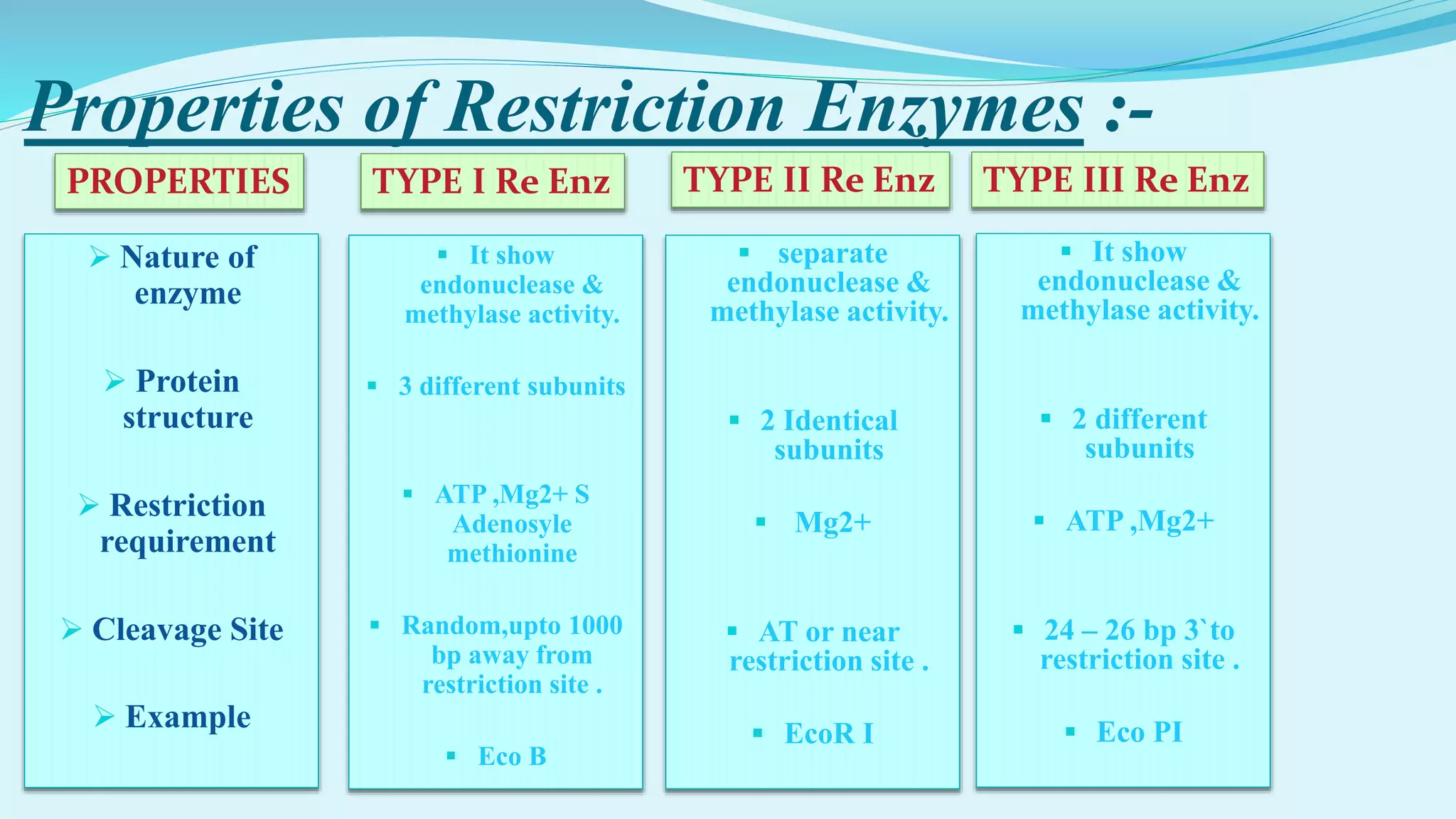
This document discusses restriction enzymes, including their discovery, types, subunits, nomenclature, recognition sequences, properties, and applications. Restriction enzymes are bacterial enzymes that cut DNA at specific recognition sequences. There are three main types - Type I cut DNA randomly, Type II cut within or near their recognition sequences, and Type III cut nearby. They are used in gene cloning, protein expression, DNA manipulation, and studying DNA sequences.