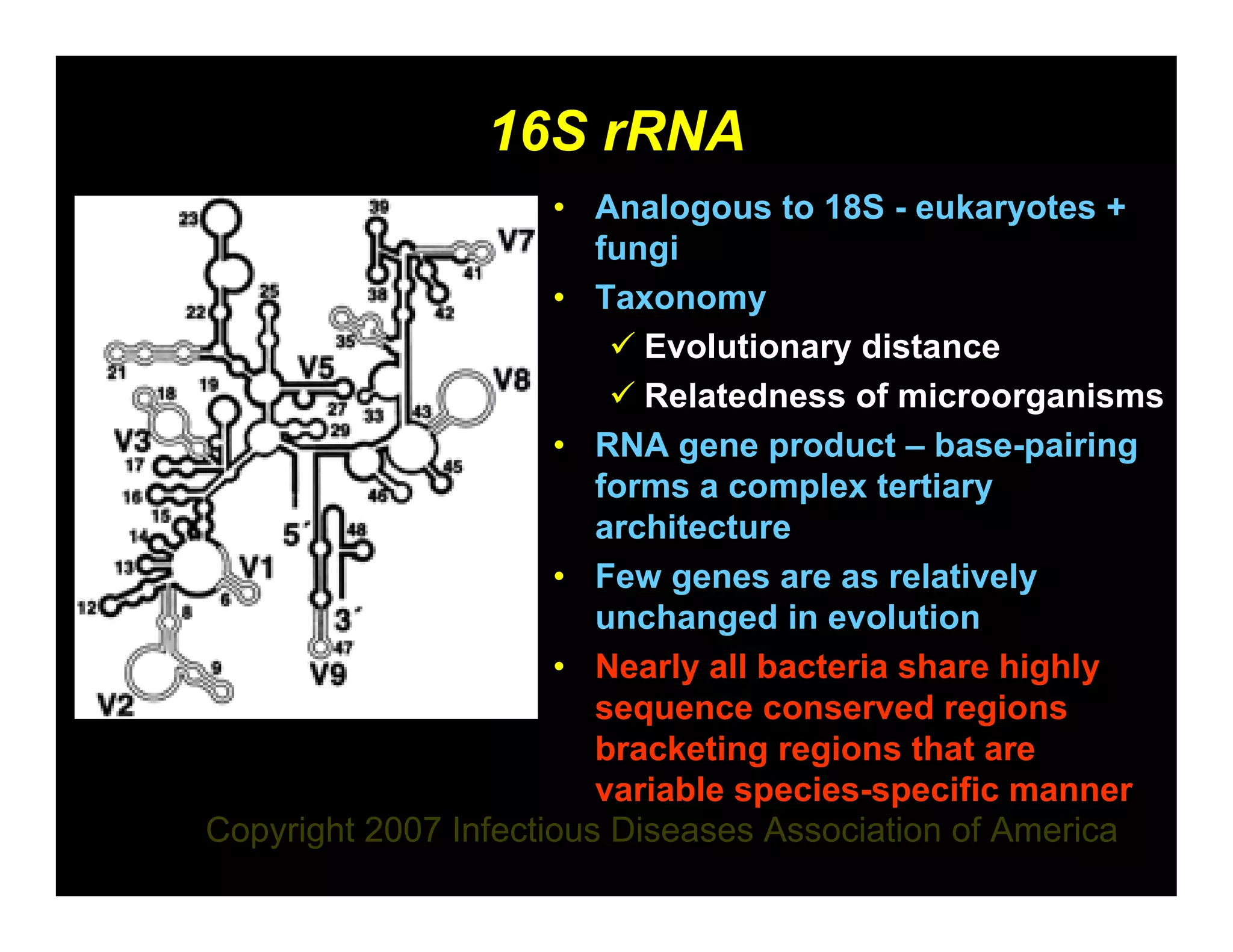
1) 16S rRNA sequencing is the gold standard for bacterial identification and can identify novel, rare, or aberrant bacterial strains that other phenotypic methods cannot.
2) The document presents data from sequencing 300 clinical isolates, identifying many new species and some new genera, and providing definitive identifications in 88% of cases.
3) While powerful, 16S rRNA sequencing has some limitations like requiring a pure culture and difficulty differentiating closely related species, and interpretation requires consideration of database issues.