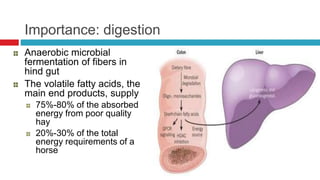
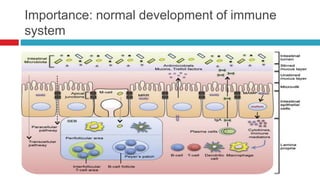
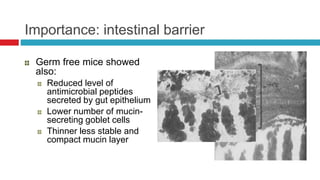
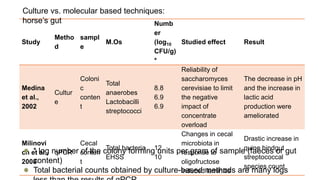
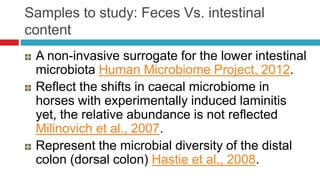
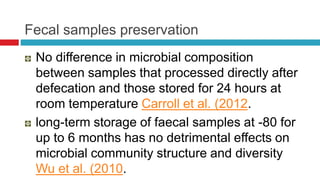
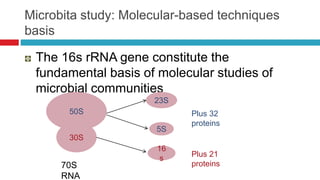
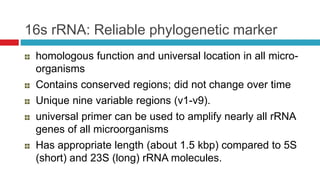
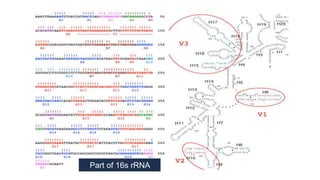
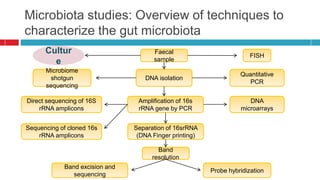
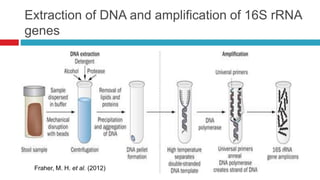
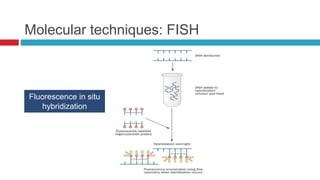
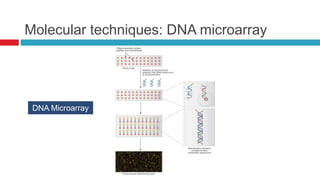
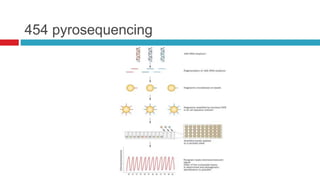
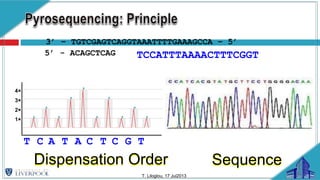
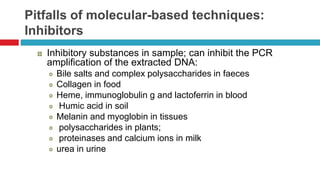
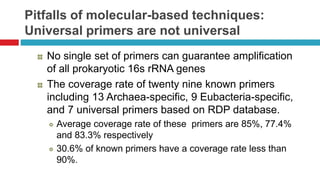
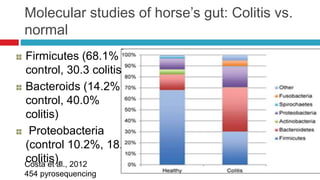
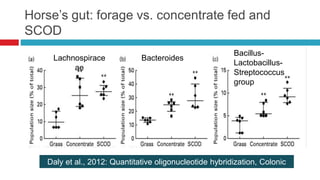
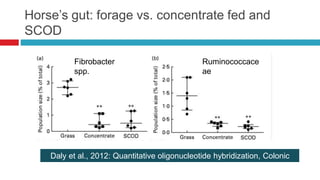
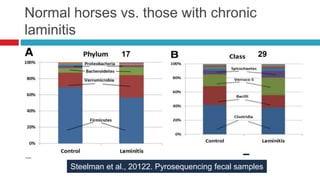
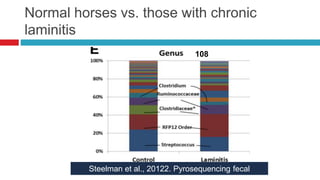
The document discusses gut microbiota and various techniques used to study it. It begins by describing the importance of gut microbiota in digestion, development of the immune system, maintenance of the intestinal barrier, and health and disease. It then covers culture-based and molecular-based techniques used to characterize gut microbiota, including their pros and cons. Specific molecular techniques discussed include FISH, DNA microarrays, DGGE/TGGE, 454 pyrosequencing, and quantitative PCR. The document also addresses pitfalls of molecular techniques and provides examples of studies comparing the gut microbiota of normal vs. diseased horses.