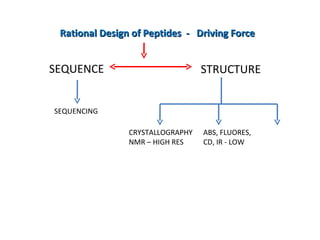
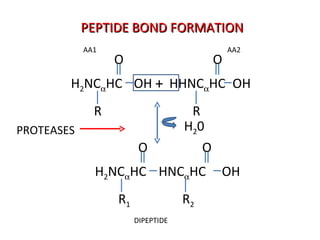
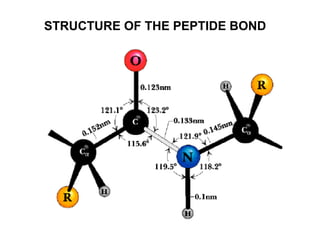
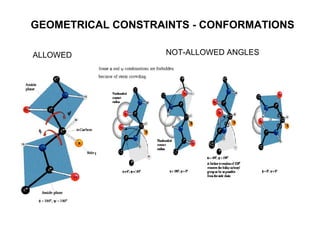
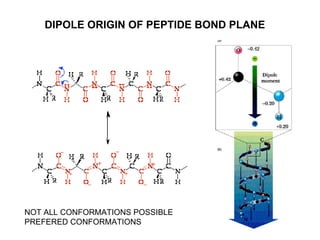
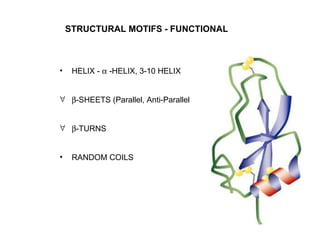
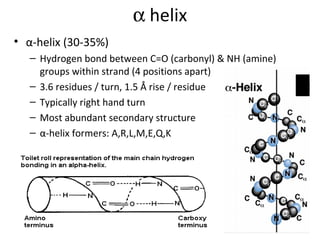
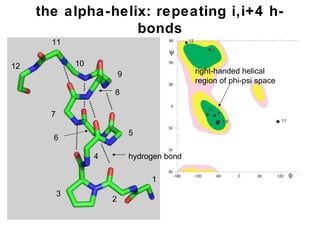
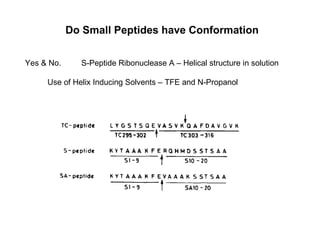
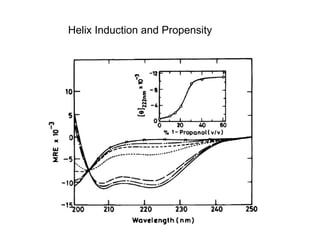
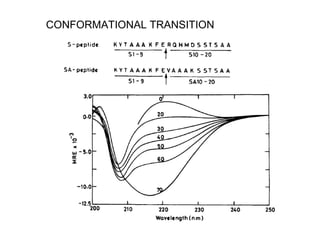
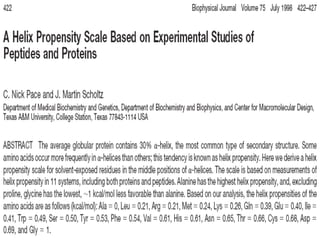
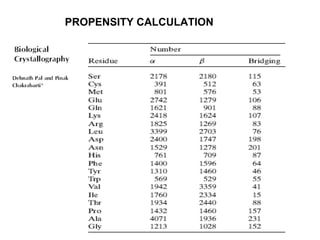
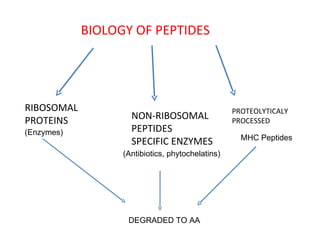
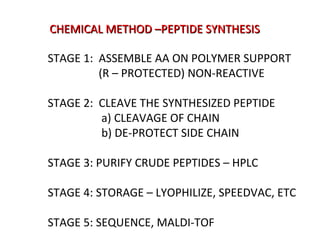
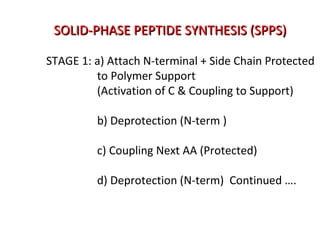
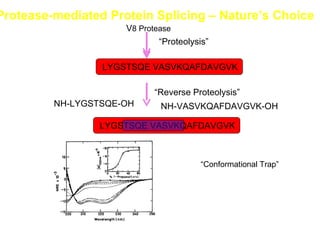
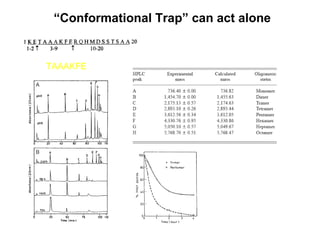
The document discusses various aspects of peptide structure and function including:
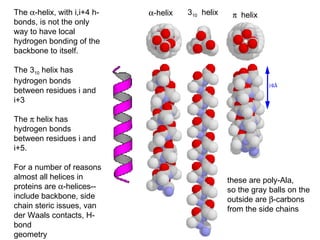
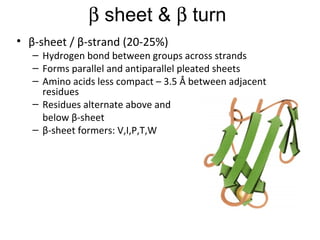
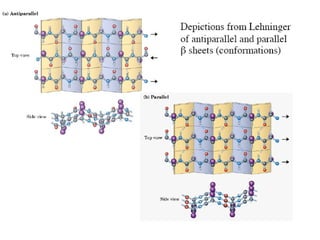
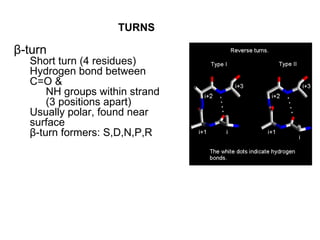
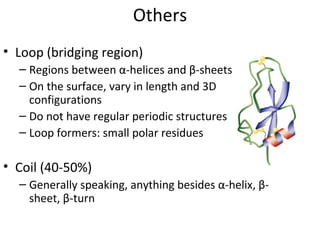
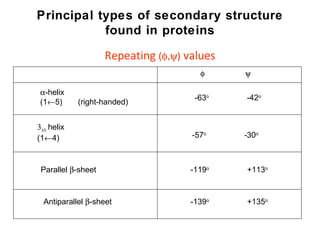
1. The common secondary structural motifs of peptides including alpha helices, beta sheets, beta turns, and random coils.
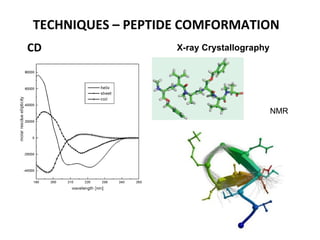
2. Techniques used to study peptide conformation such as crystallography, NMR, and CD spectroscopy.
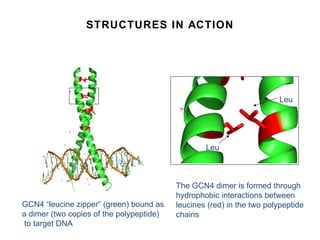
3. Factors that influence peptide structure including amino acid sequence, solvent environment, and intermolecular interactions.