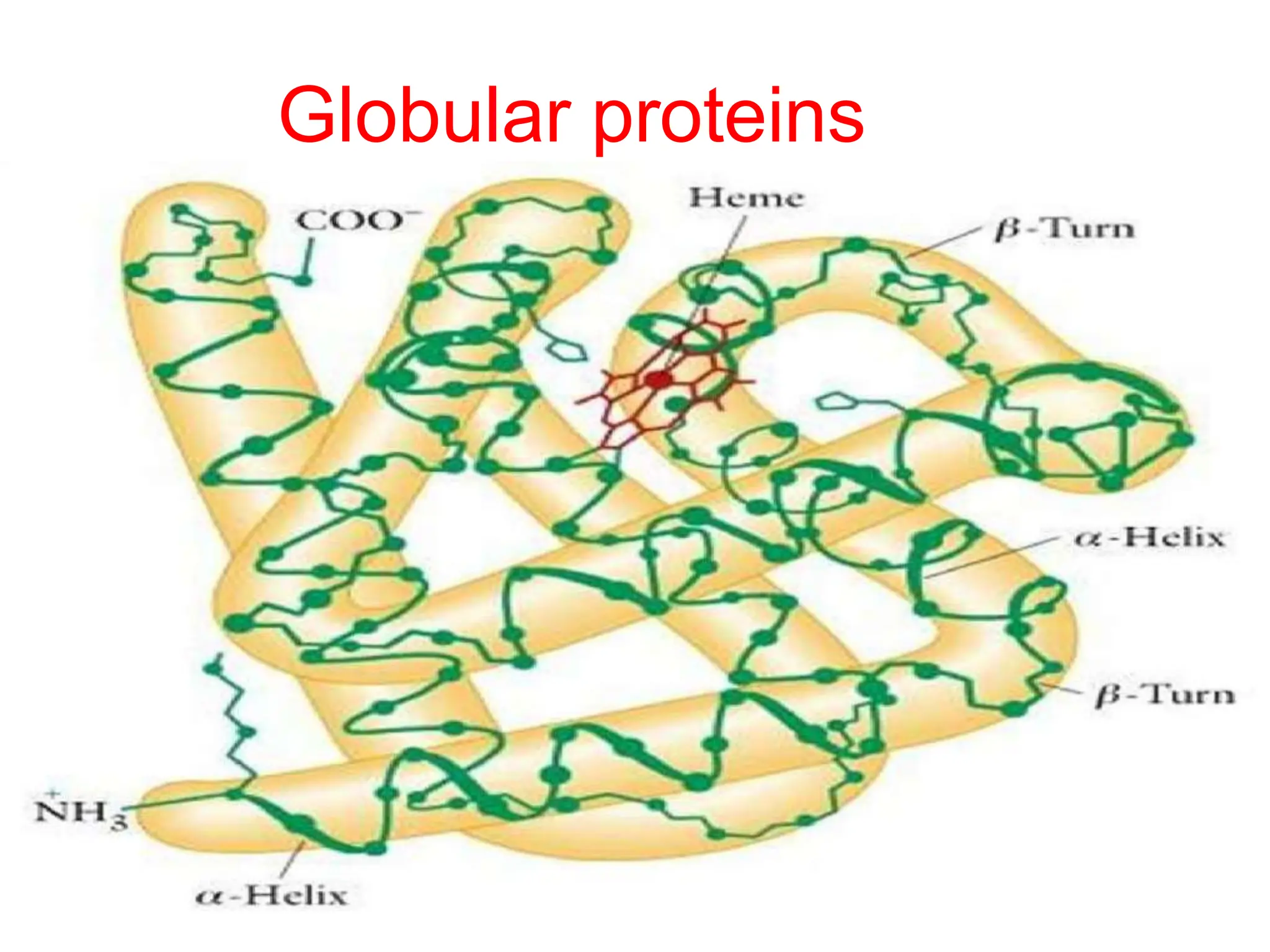
The document provides a detailed overview of protein structure and function, highlighting that proteins are essential organic molecules made of amino acids. It explains the levels of organization in proteins, including primary, secondary, tertiary, and quaternary structures, and discusses the various interactions that stabilize these structures. Additionally, it contrasts globular proteins, which are compact and versatile, with fibrous proteins that serve structural roles.