1. The document discusses various methods for quantifying two-dimensional shapes and textures in medical images, including statistical moments, spatial moments, radial distance measures, chain codes, Fourier descriptors, thinning, and texture measures.
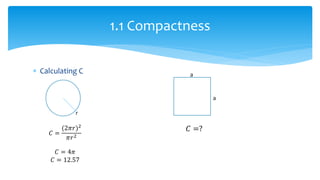
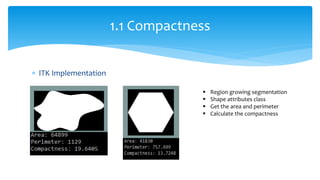
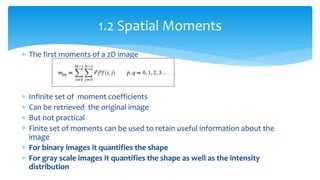
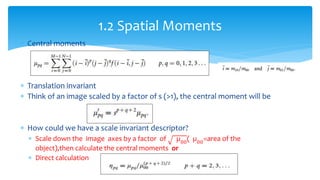
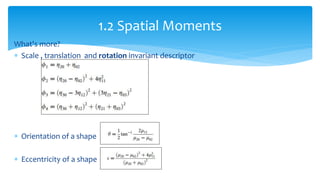
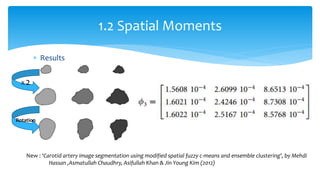
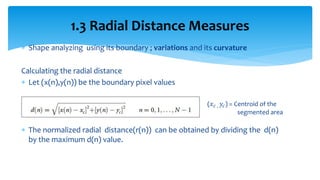
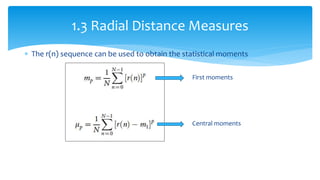
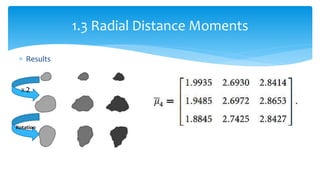
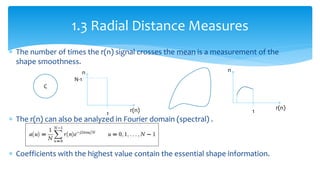
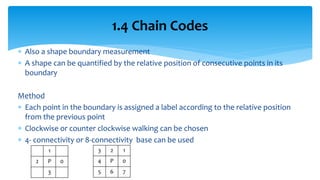
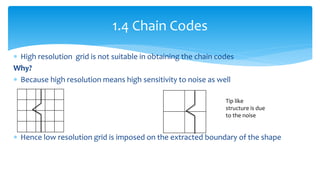
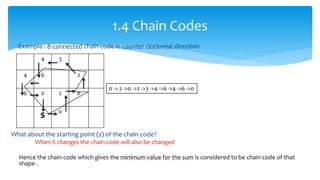
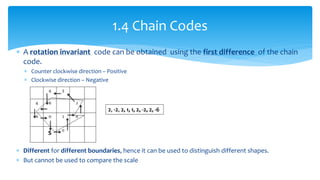
2. Compactness, calculated using perimeter and area, quantifies how close a shape is to a circle. Spatial moments provide quantitative measurements of point distributions and shapes. Radial distance measures analyze boundary curvature. Chain codes represent boundary points.
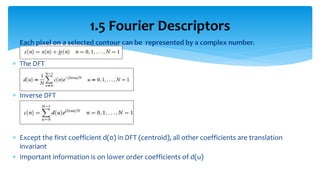
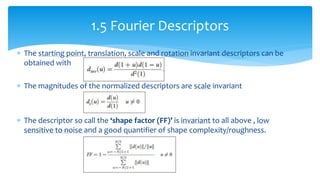
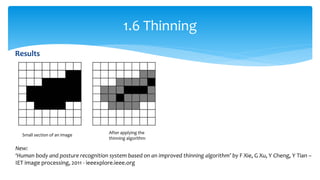
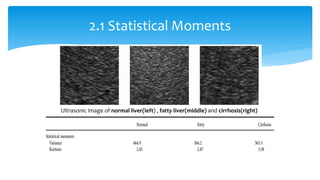
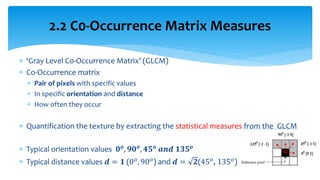
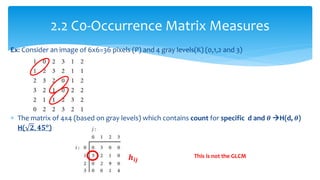
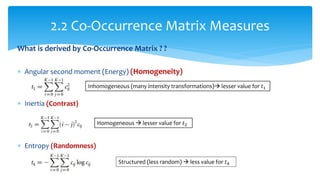
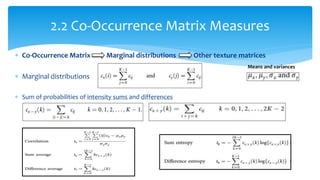
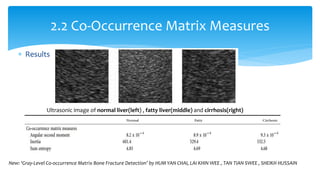
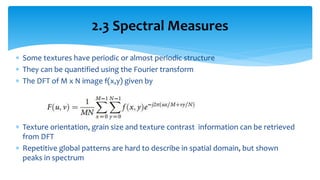
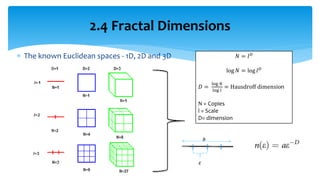
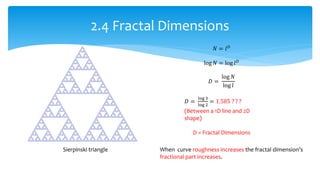
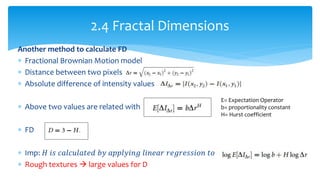
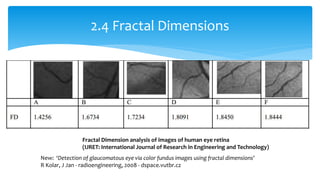
3. Fourier descriptors and thinning/skeletonization reduce shapes to descriptors and graphs for analysis. Texture is quantified using statistical moments, co-occurrence matrices, spectral measures, and fractal dimensions.