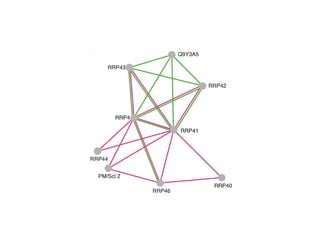
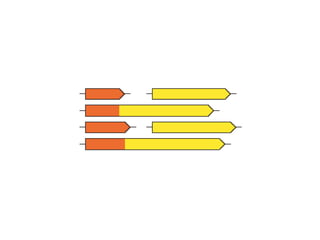
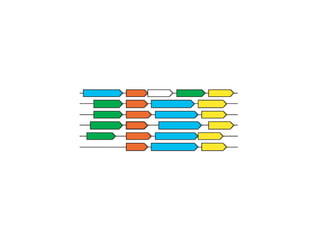
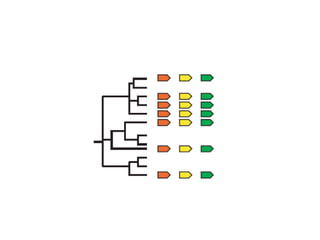
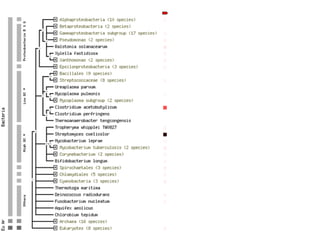
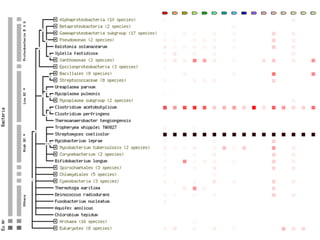
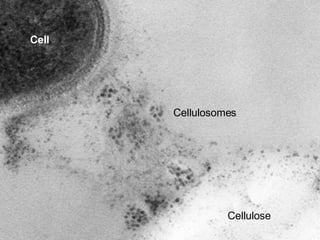
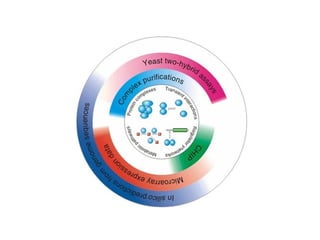
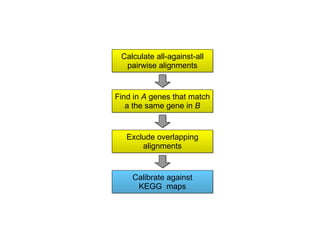
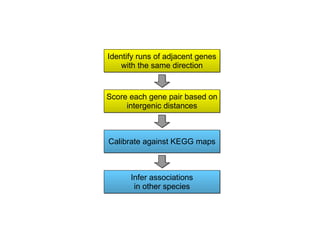
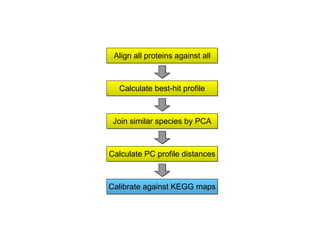
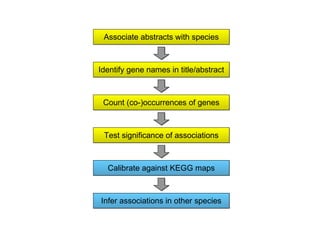
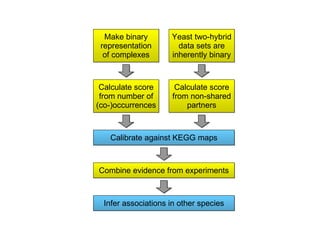
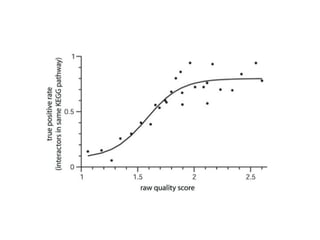
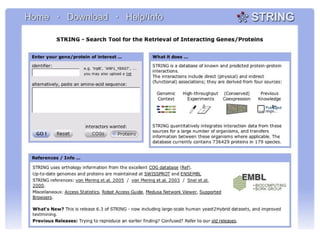
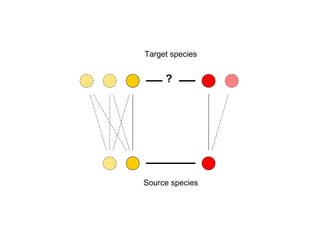
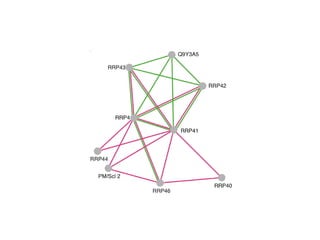
STRING integrates diverse evidence about functional interactions between proteins from hundreds of proteomes. It combines data from genomic context methods, curated databases, experiments, and textmining to generate a global network of protein interactions. The different evidence sources have issues like inconsistent identifiers, variable quality, and coverage of different species that STRING addresses through parsers, orthology transfer, and quality scores to generate a single confidence score for each interaction.