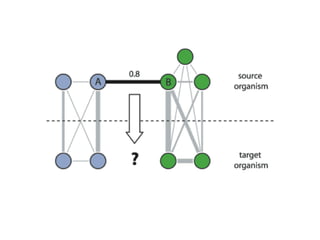
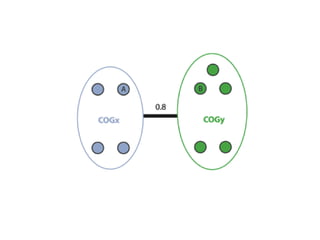
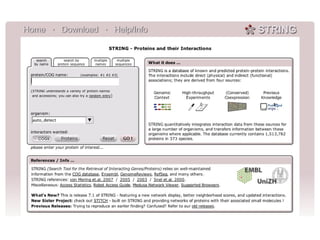
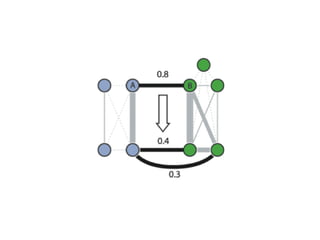
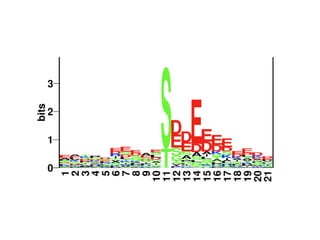
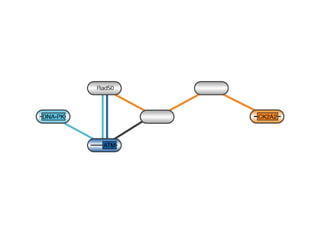
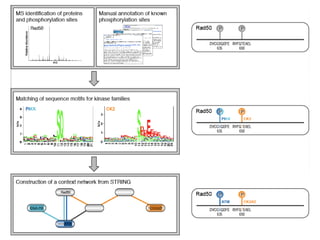
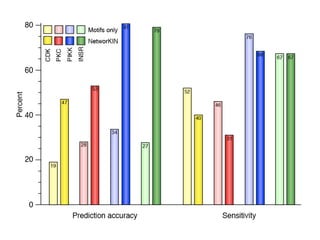
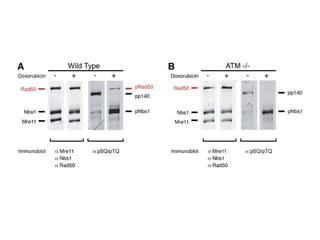
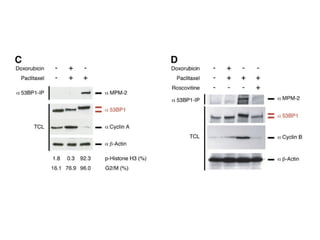
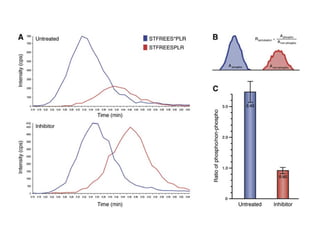
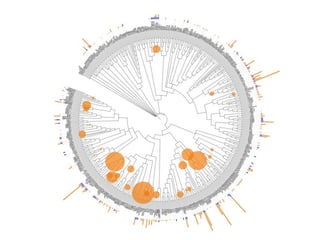
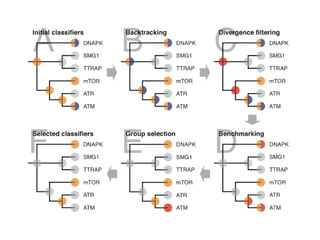
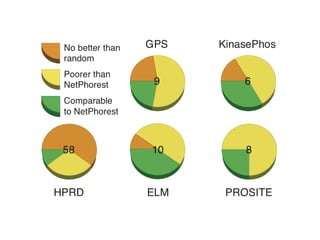
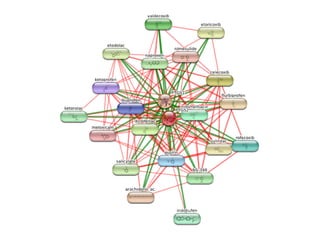
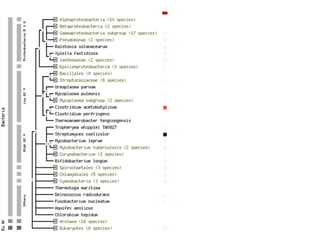
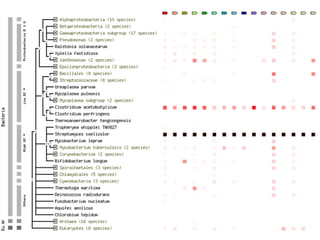
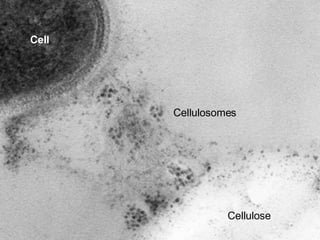
The document discusses the integration of heterogeneous data in biological research, emphasizing methodologies like phylogenetic profiling, gene expression analysis, and interaction networks. It highlights various resources and databases such as STRING, Biogrid, and Netphorest for data mining and analysis of gene and protein interactions. Additionally, it addresses challenges like data quality, context localization, and the need for computational validation methods in signaling networks.

































![Gene and protein names Cue words for entity recognition Verbs for relation extraction [ nxexpr T he expression of [ nxgene the cytochrome genes [ nxpg CYC1 and CYC7 ]]] is controlled by [ nxpg HAP1 ]](https://image.slidesharecdn.com/jen08talk3-1217978434911536-9/85/Network-integration-of-heterogeneous-data-52-320.jpg)