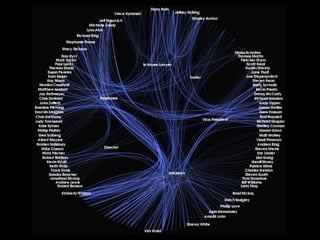
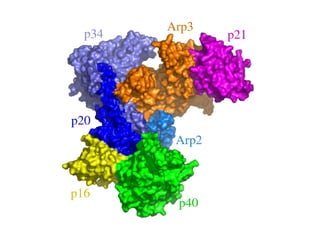
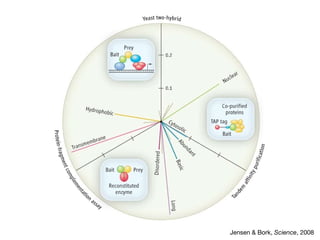
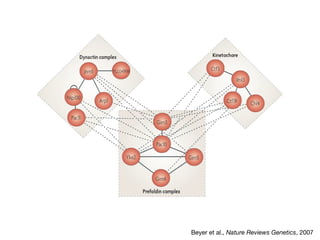
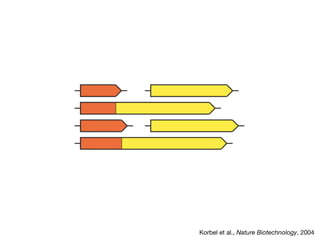
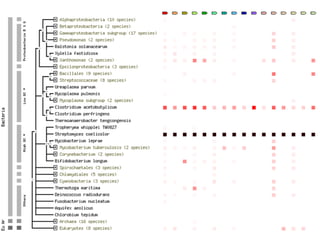
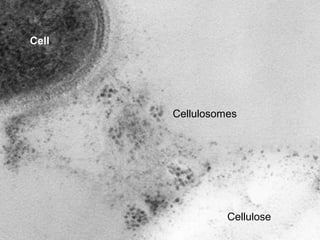
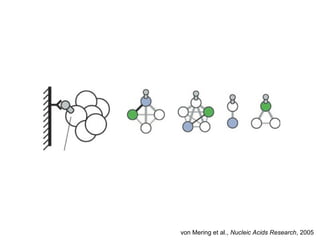
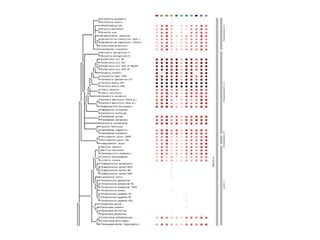
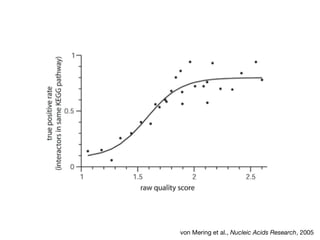
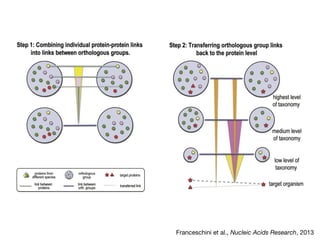
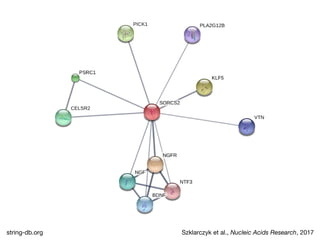
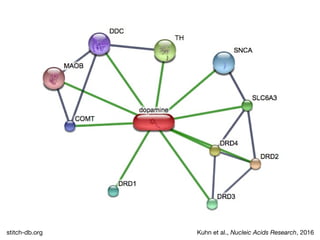
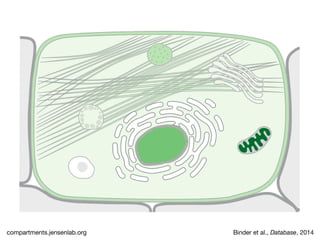
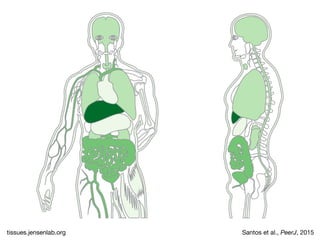
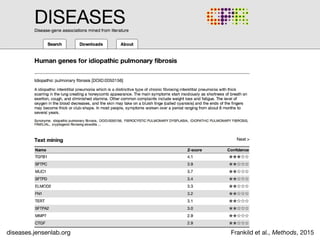
The document discusses the STRING database, which integrates heterogeneous biological data to generate association networks for proteins. It describes how STRING collects and connects curated knowledge, experimental data, and predicted interactions from genomic context, co-expression and text mining. The document also outlines exercises for users to explore protein-protein associations in STRING and related databases that integrate data on subcellular localization, tissue expression, and disease associations.