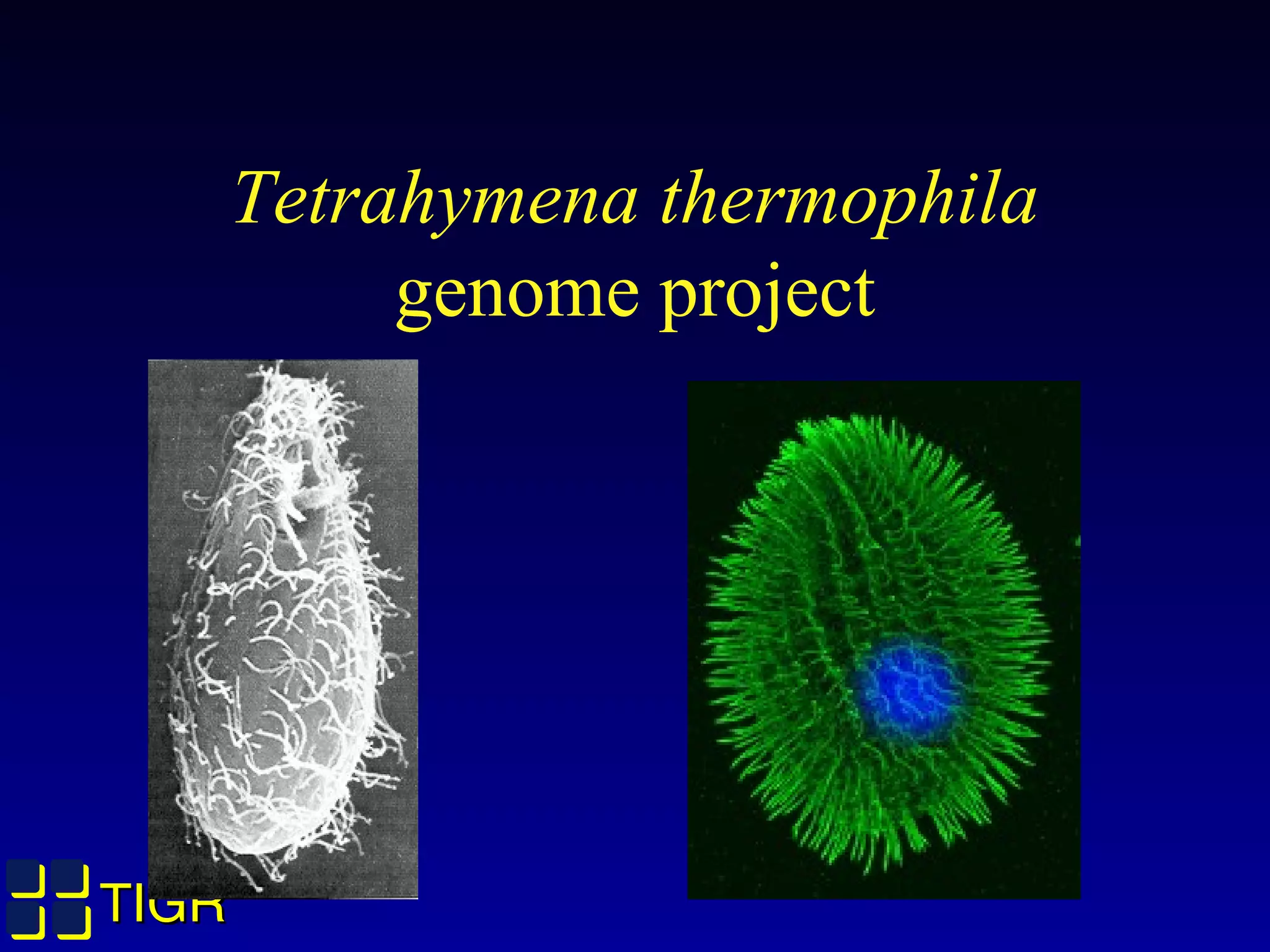
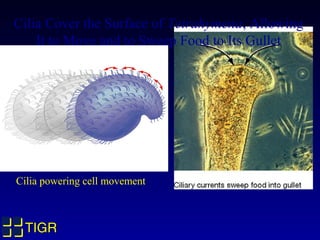
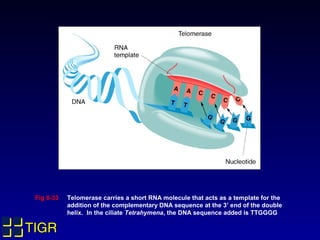
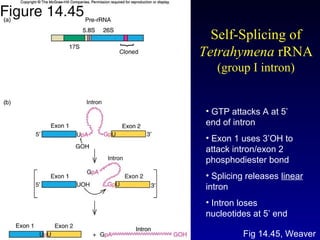
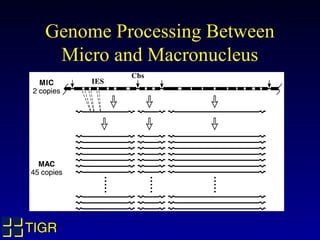
The Tetrahymena thermophila Genome Project, coordinated by Ed Orias, aims to explore the genomic and biological features of this freshwater ciliate, which serves as a model organism for eukaryotic studies. Major discoveries linked to Tetrahymena include insights into RNA catalysis, telomeres, and cellular processes, supported by advanced genetic tools and a robust research community. The project encompasses genome mapping, gene annotation, and the development of bioinformatics resources to facilitate research on gene knockouts and cellular functions.