This document provides a project update on Biopython. Biopython is a freely available Python library for biological computation maintained by contributors in a distributed collaboration. The update summarizes new features in Biopython including improved graphics capabilities, support for additional file formats, integration with external tools like PAML, and two ongoing Google Summer of Code projects involving genomic variants and search results input/output.

![Hello, BOSC
Biopython is a freely available Python library for biological
computation, and a long-running, distributed collaboration
to produce and maintain it [1].
● Supported by the Open Bioinformatics Foundation
(OBF)
● "This is Python's Bio* library. There are several Bio*
libraries like it, but this one is ours."
● http://biopython.org/
_____
[1] Cock, P.J.A., Antao, T., Chang, J.T., Chapman, B.A., Cox, C.J., Dalke, A.,
Friedberg, I., Hamelryck, T., Kauff, F., Wilczynski, B., de Hoon, M.J. (2009)
Biopython: freely available Python tools for computational molecular biology
and bioinformatics. Bioinformatics 25(11) 1422-3. doi:10.1093
/bioinformatics/btp163](https://image.slidesharecdn.com/biopython-project-update-120714154824-phpapp02/85/E-Talevich-Biopython-project-update-2-320.jpg)
![Bio.Graphics (Biopython 1.59, February 2012)
New features in...
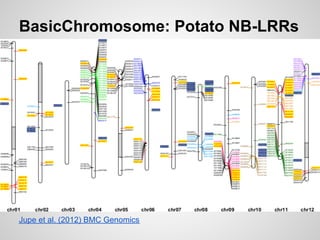
BasicChromosome:
● Draw simple sub-features on chromosome segments
● Show the position of genes, SNPs or other loci
GenomeDiagram [2]:
● Cross-links between tracks
● Track-specific start/end positions for showing regions
_____
[2] Pritchard, L., White, J.A., Birch, P.R., Toth, I. (2010) GenomeDiagram: a
python package for the visualization of large-scale genomic data.
Bioinformatics 2(5) 616-7.
doi:10.1093/bioinformatics/btk021](https://image.slidesharecdn.com/biopython-project-update-120714154824-phpapp02/85/E-Talevich-Biopython-project-update-3-320.jpg)
![SeqIO and AlignIO
(Biopython 1.58, August 2011)
● SeqXML format [3]
● Read support for ABI chromatogram files (Wibowo A.)
● "phylip-relaxed" format (Connor McCoy, Brandon I.)
○ Relaxes the 10-character limit on taxon names
○ Space-delimited instead
○ Used in RAxML, PhyML, PAML, etc.
_____
[3] Schmitt et al. (2011) SeqXML and OrthoXML: standards for sequence and
orthology information. Briefings in Bioinformatics 12(5): 485-488. doi:10.1093
/bib/bbr025](https://image.slidesharecdn.com/biopython-project-update-120714154824-phpapp02/85/E-Talevich-Biopython-project-update-7-320.jpg)
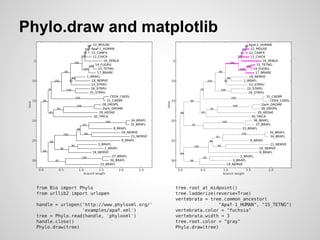
![Bio.Phylo & pypaml
● PAML interop: wrappers, I/O, glue
○ Merged Brandon Invergo’s pypaml as
Bio.Phylo.PAML (Biopython 1.58, August 2011)
● Phylo.draw improvements
● RAxML wrapper (Biopython 1.60, June 2012)
● Paper in review [4]
_____
[4] Talevich, E., Invergo, B.M., Cock, P.J.A., Chapman, B.A. (2012) Bio.Phylo:
a unified toolkit for processing, analysis and visualization of phylogenetic data
in Biopython. BMC Bioinformatics, in review](https://image.slidesharecdn.com/biopython-project-update-120714154824-phpapp02/85/E-Talevich-Biopython-project-update-8-320.jpg)