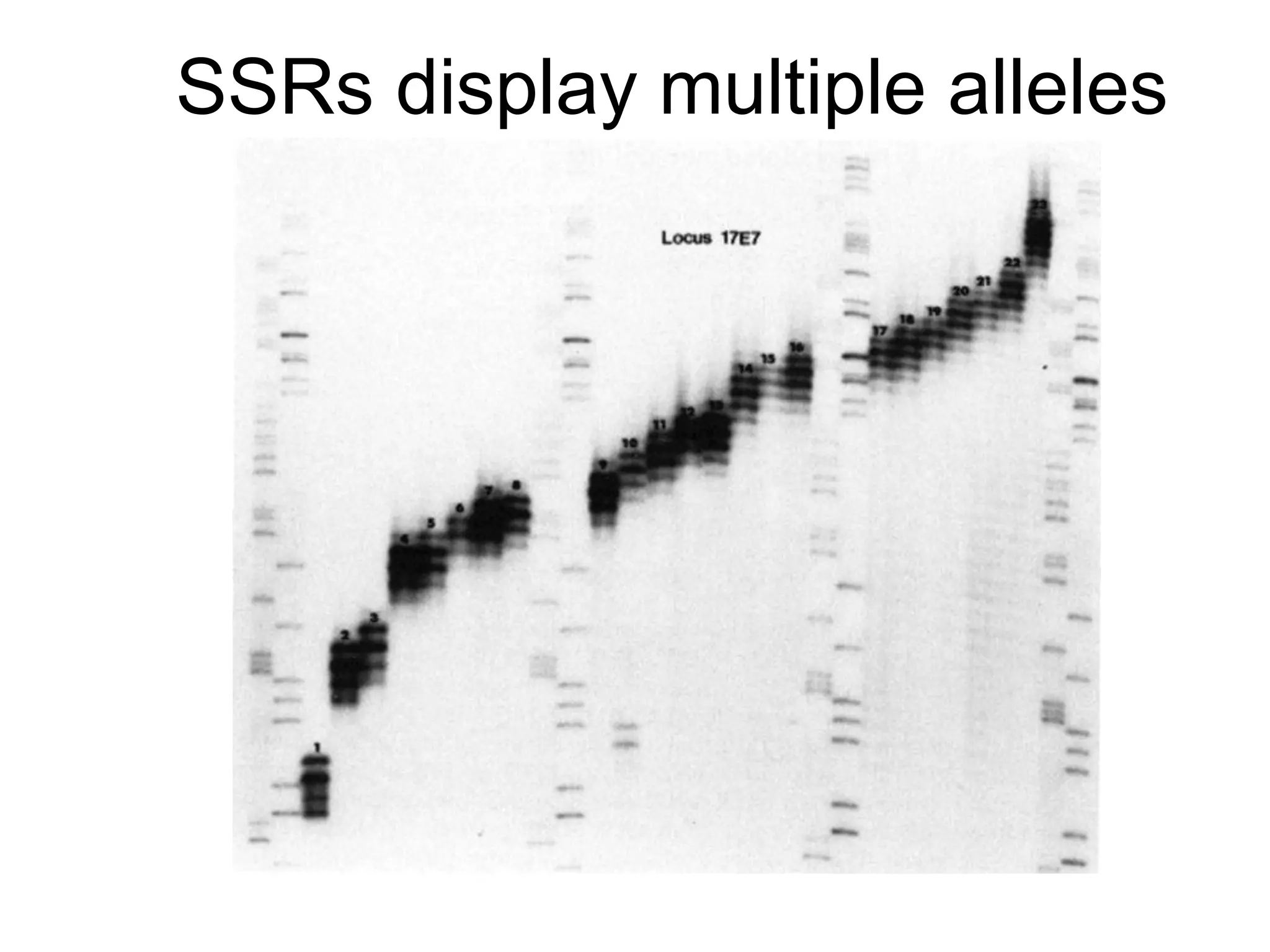
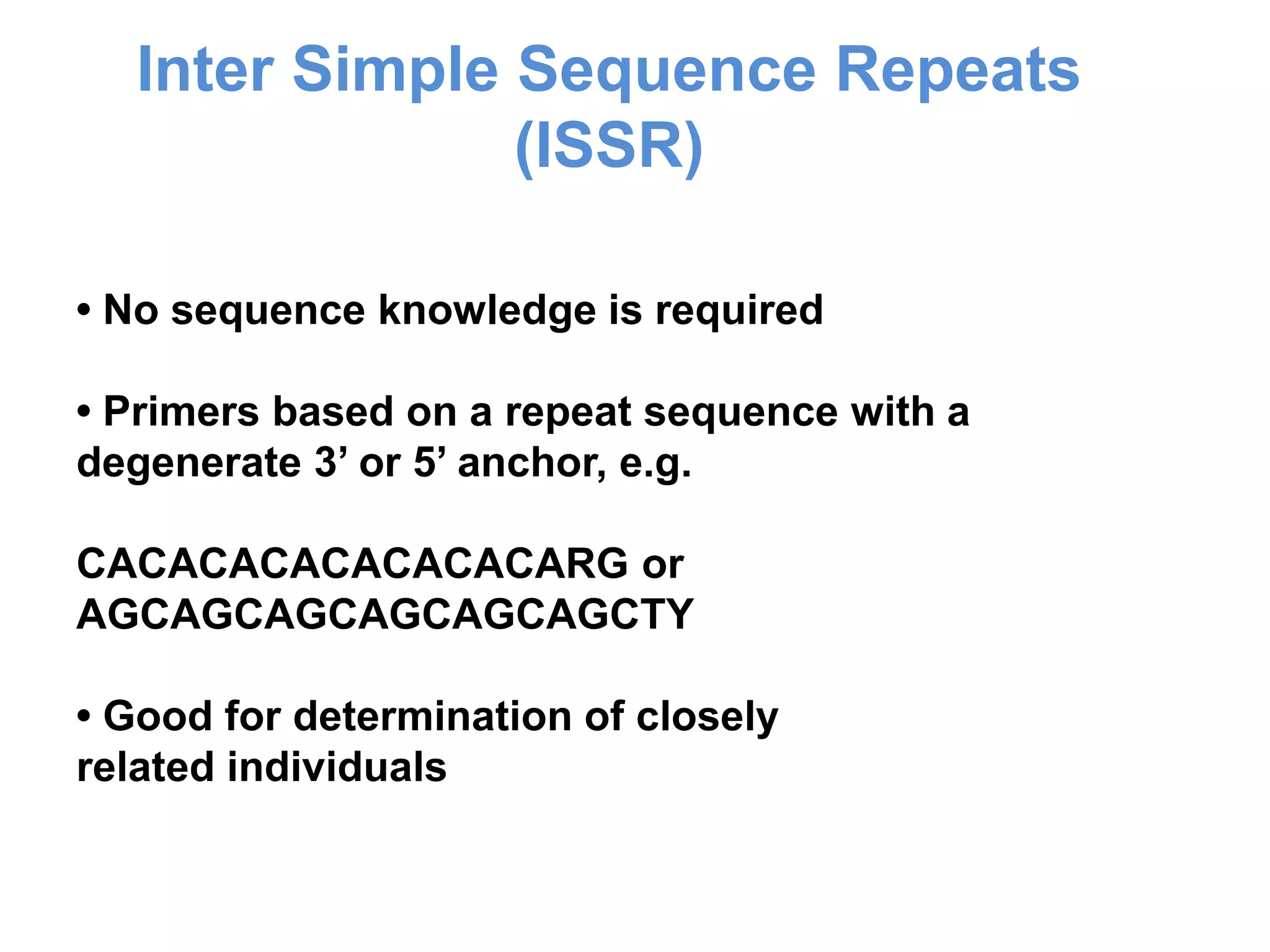
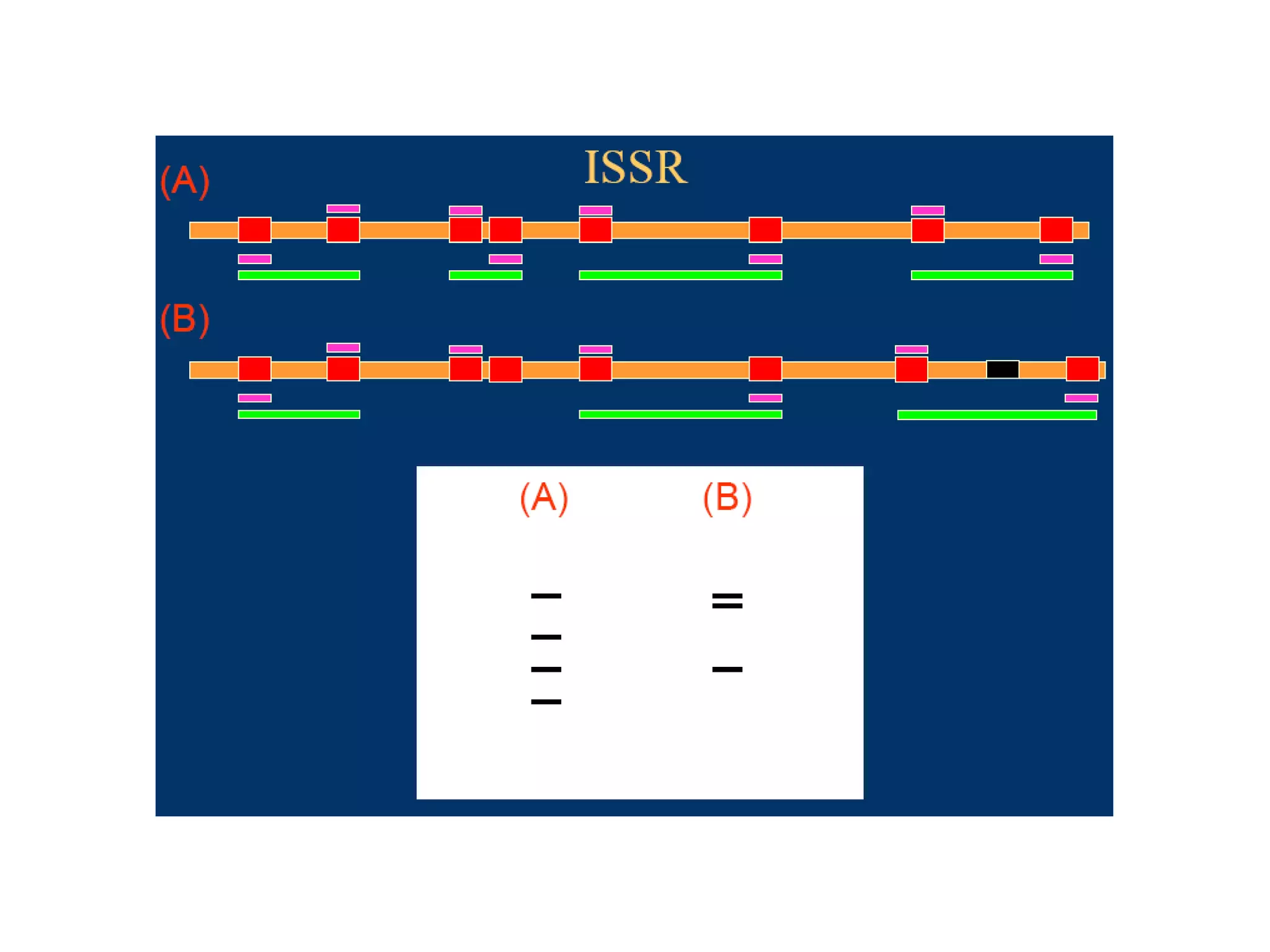
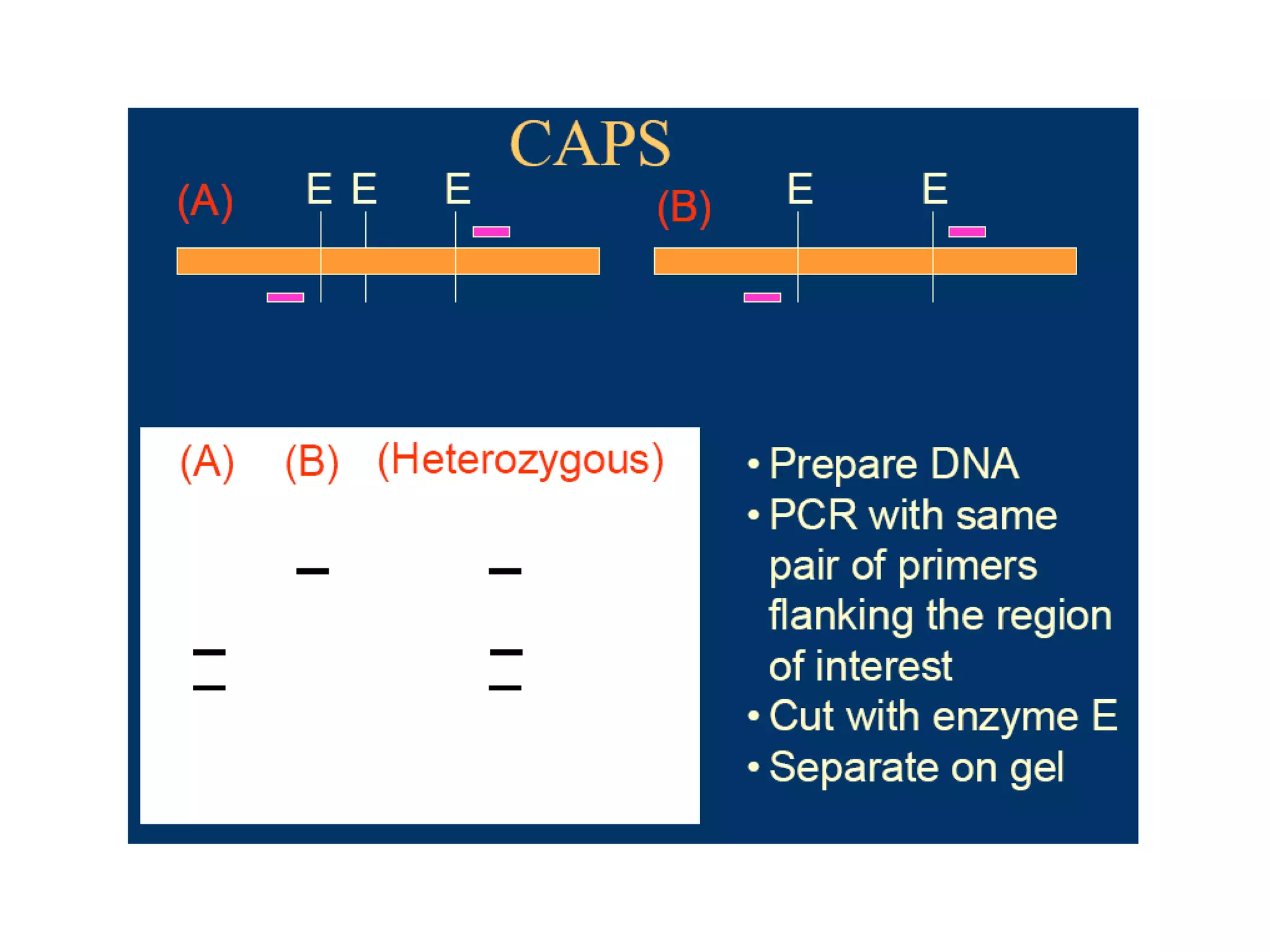
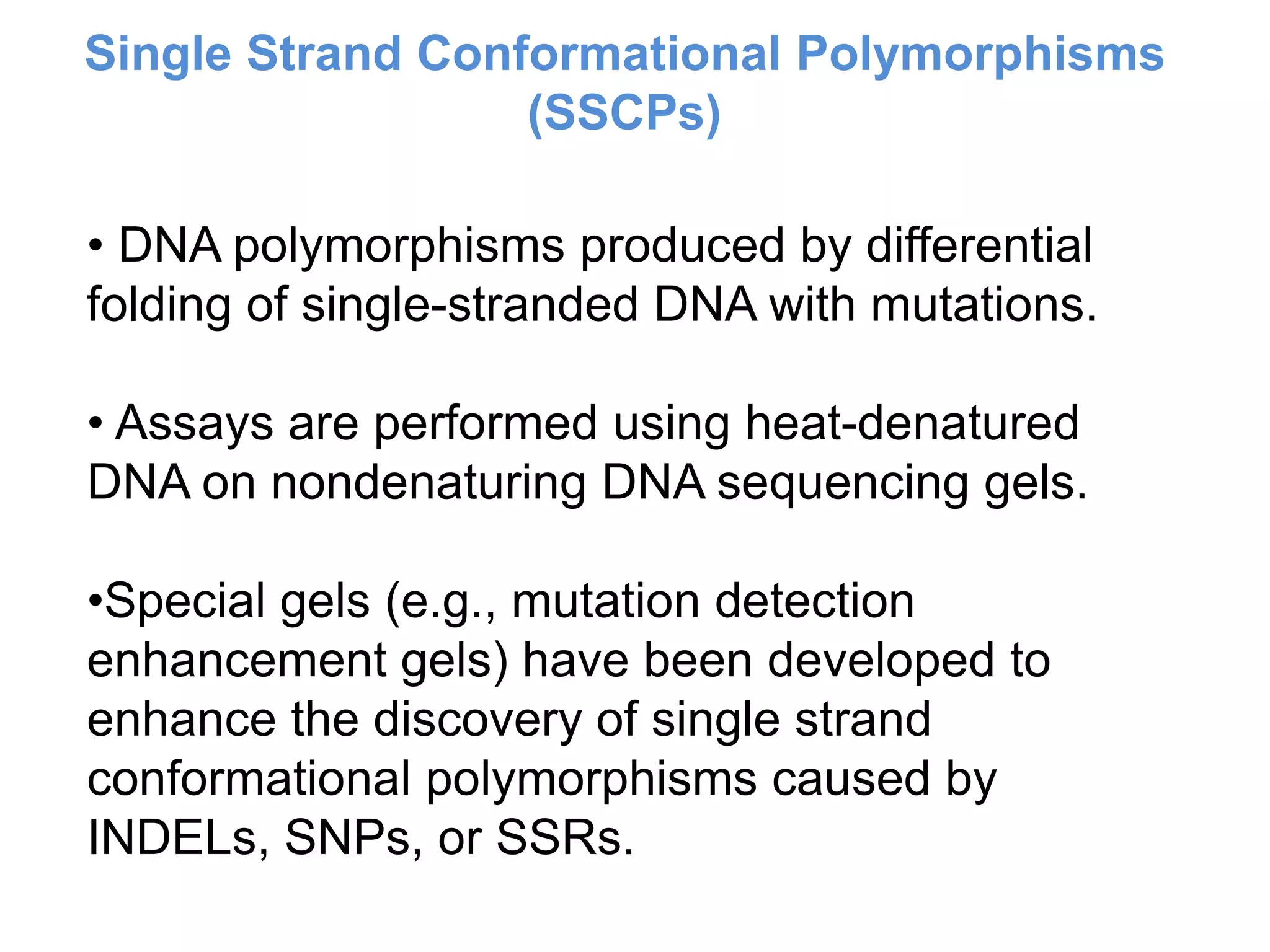
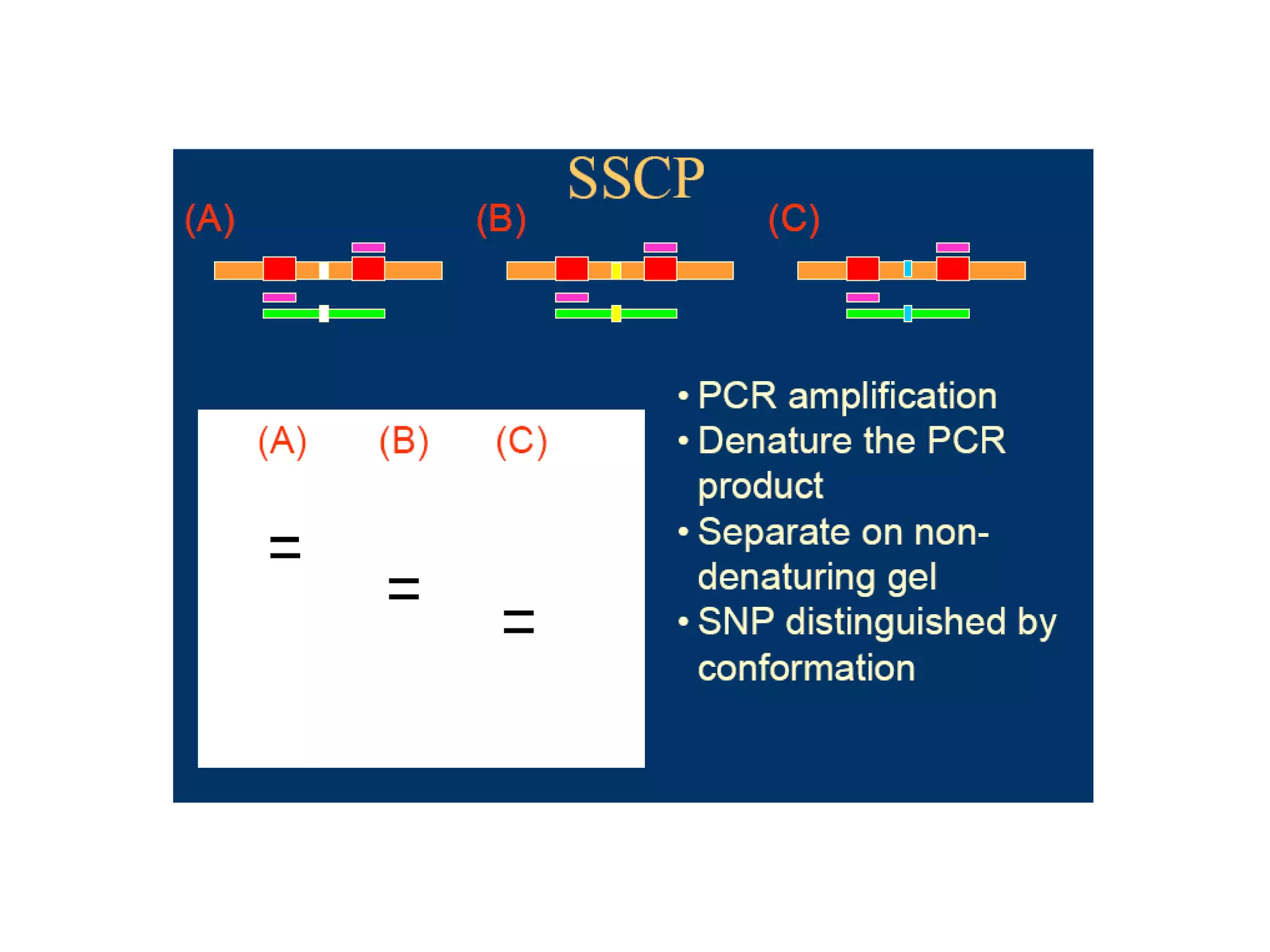
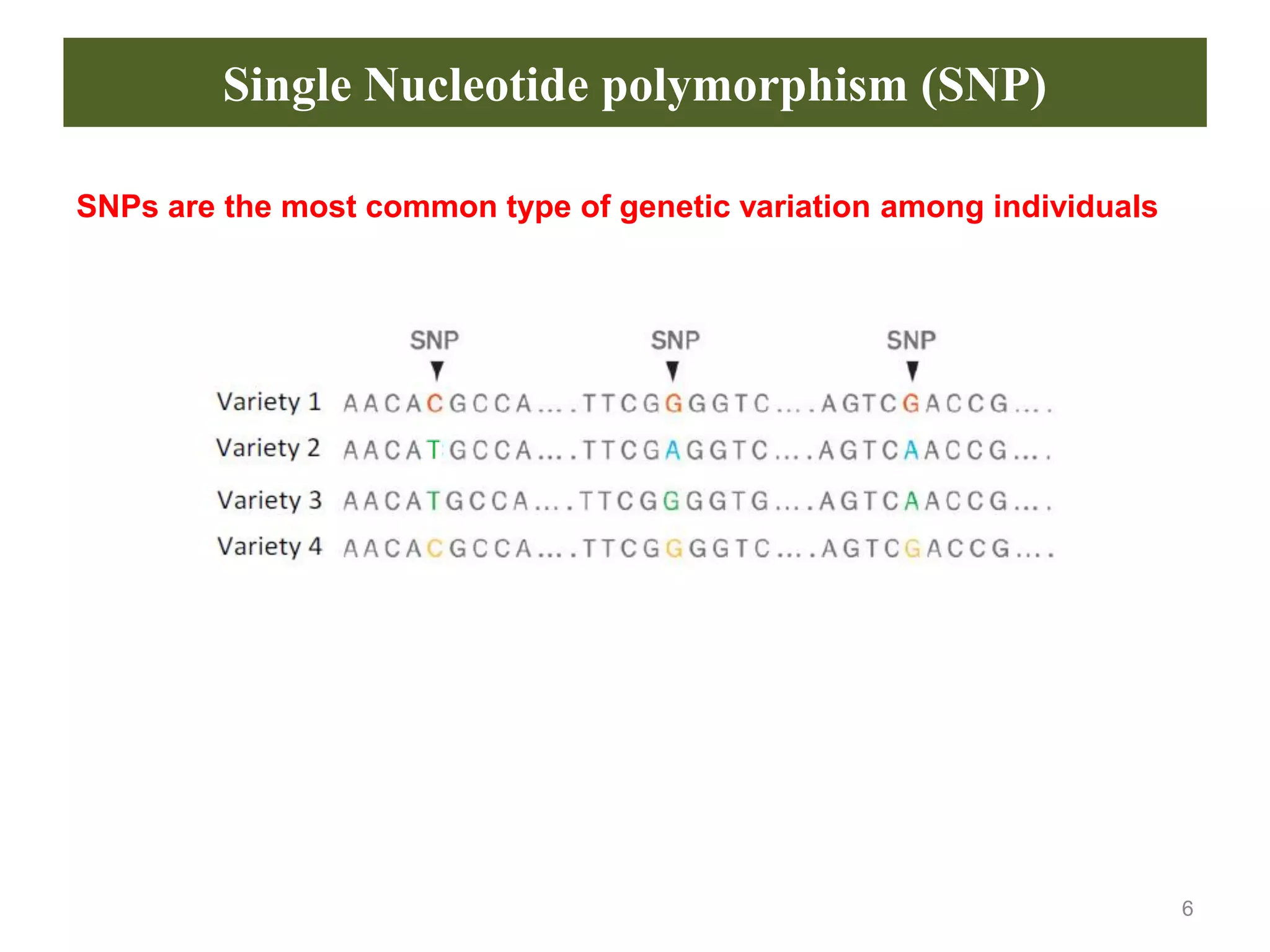
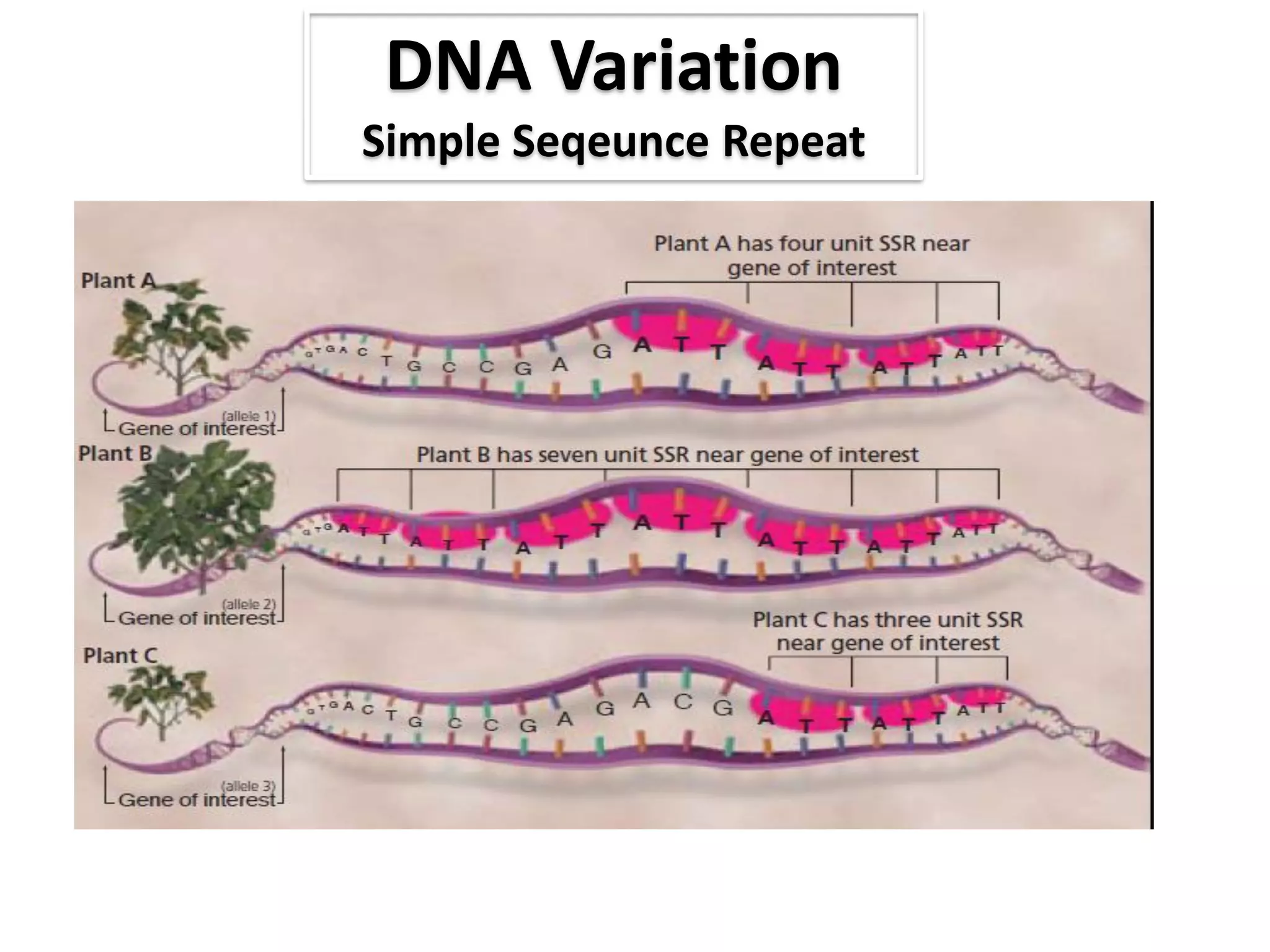
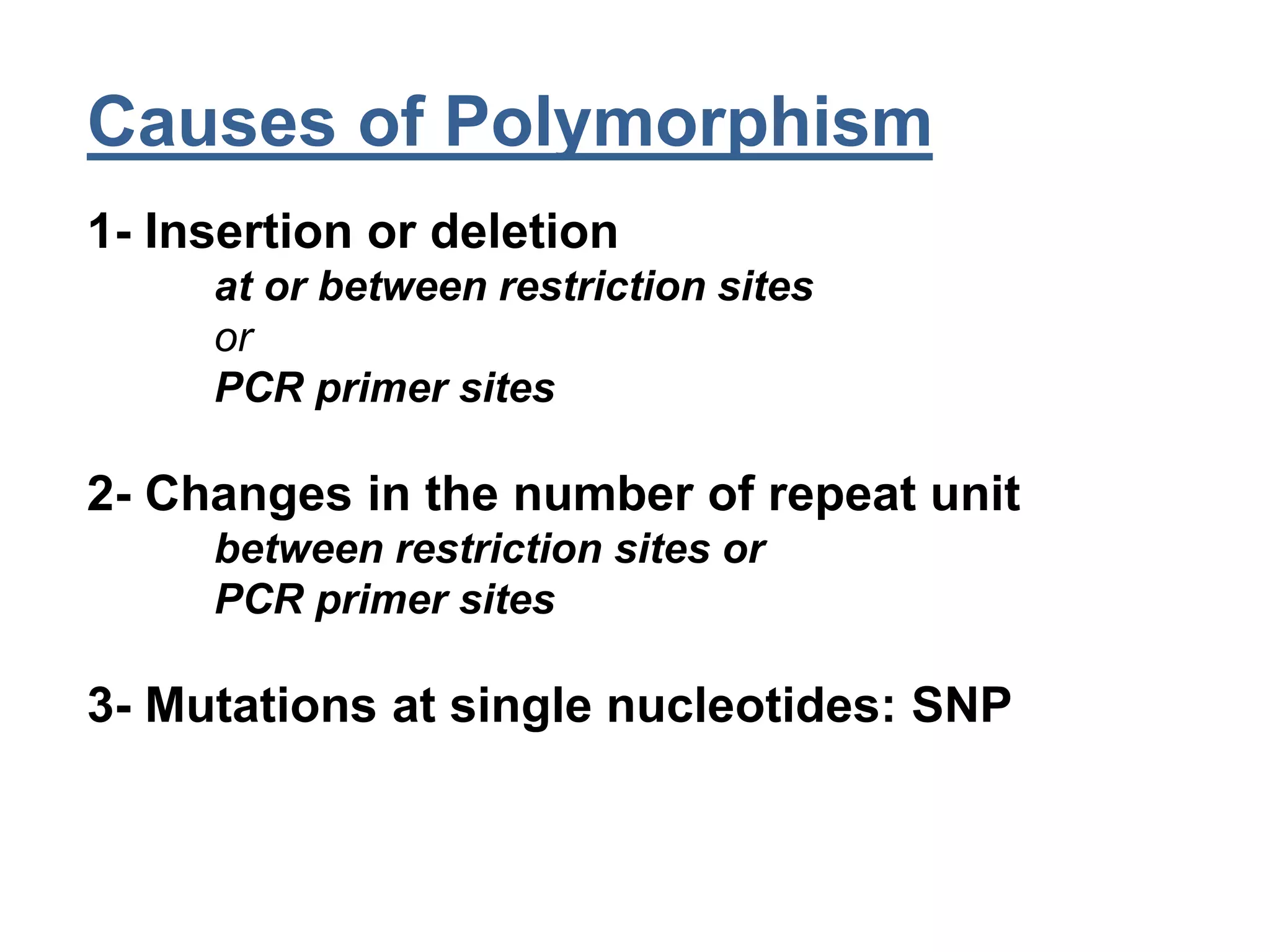
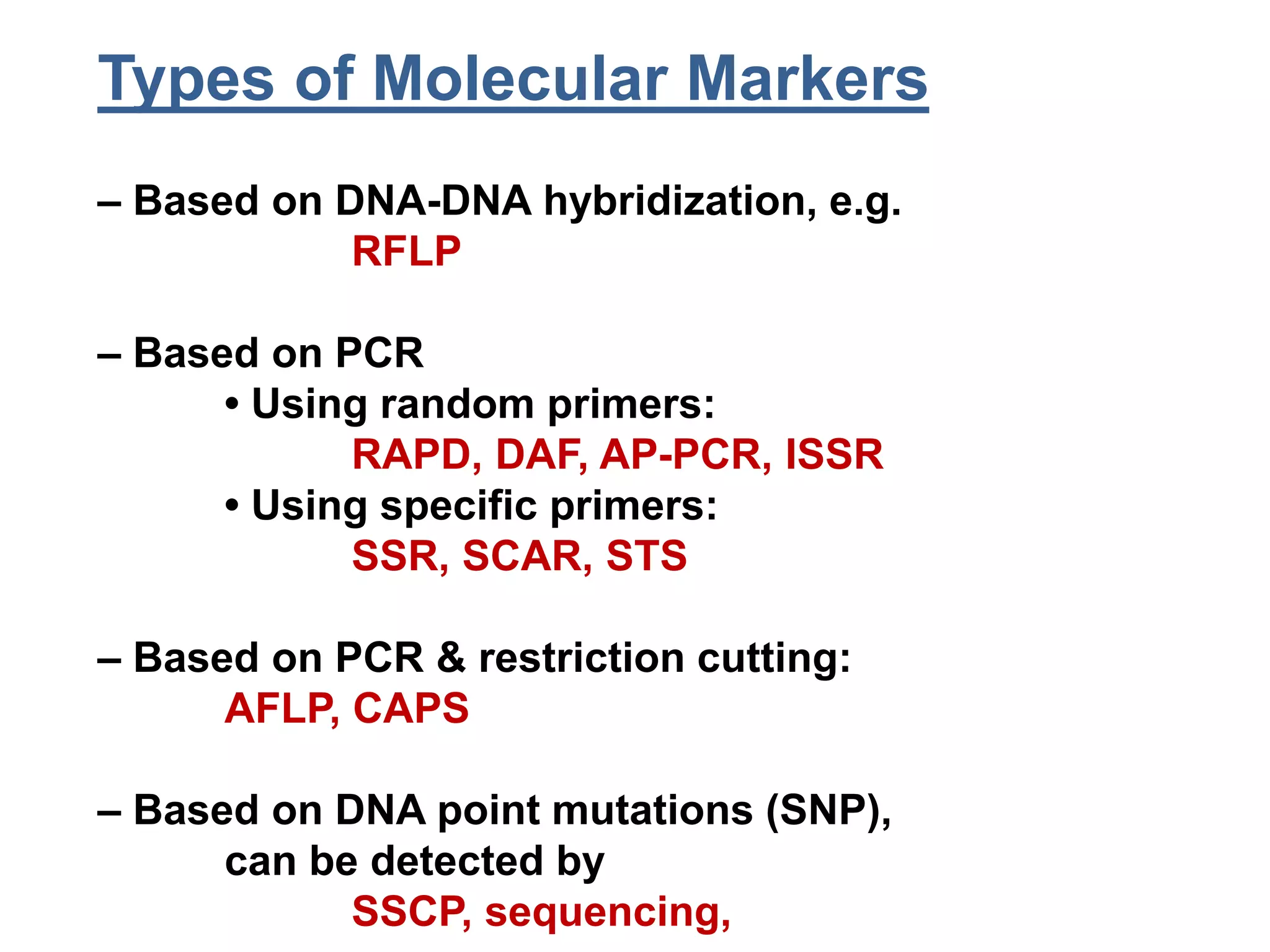
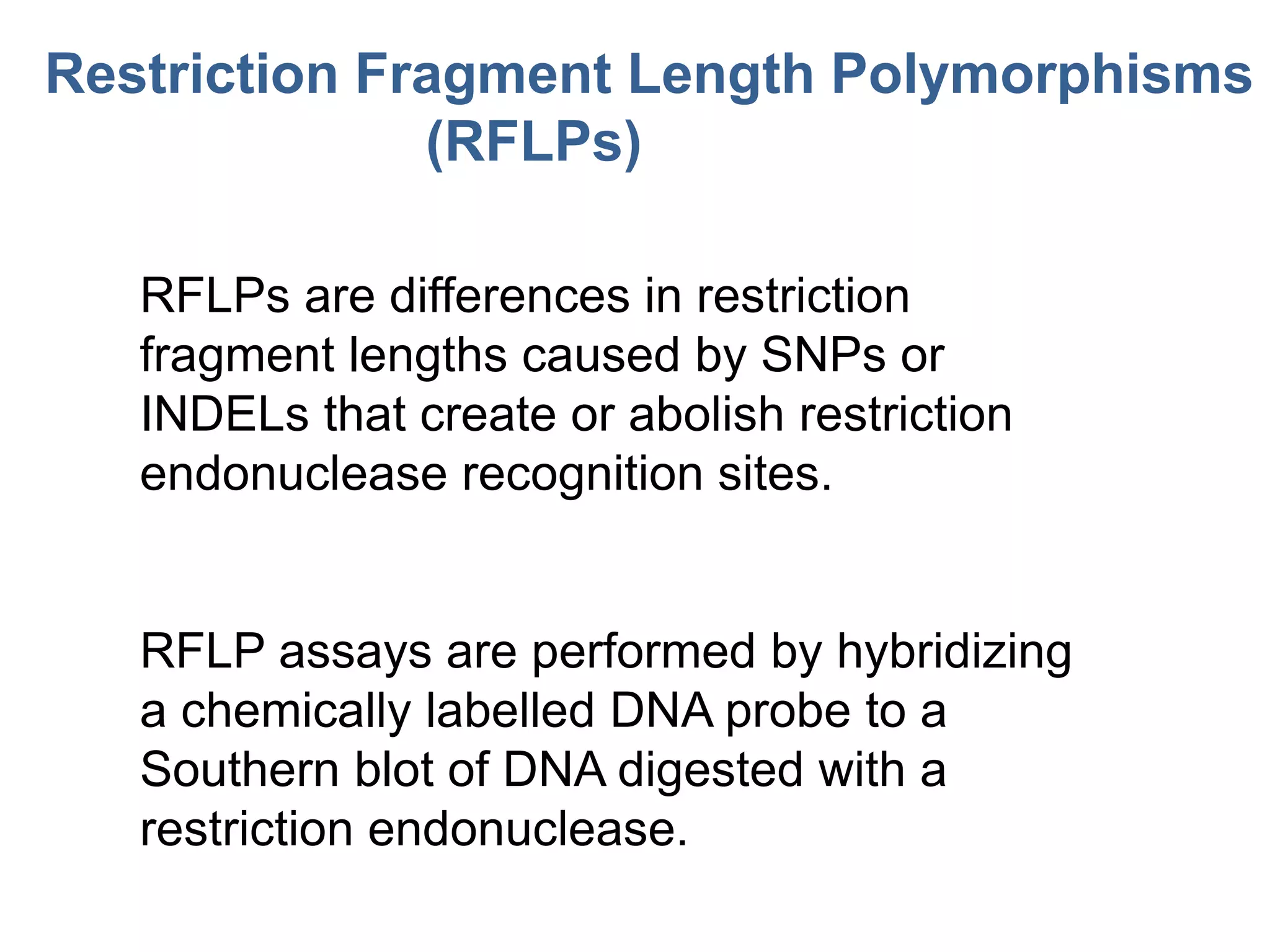
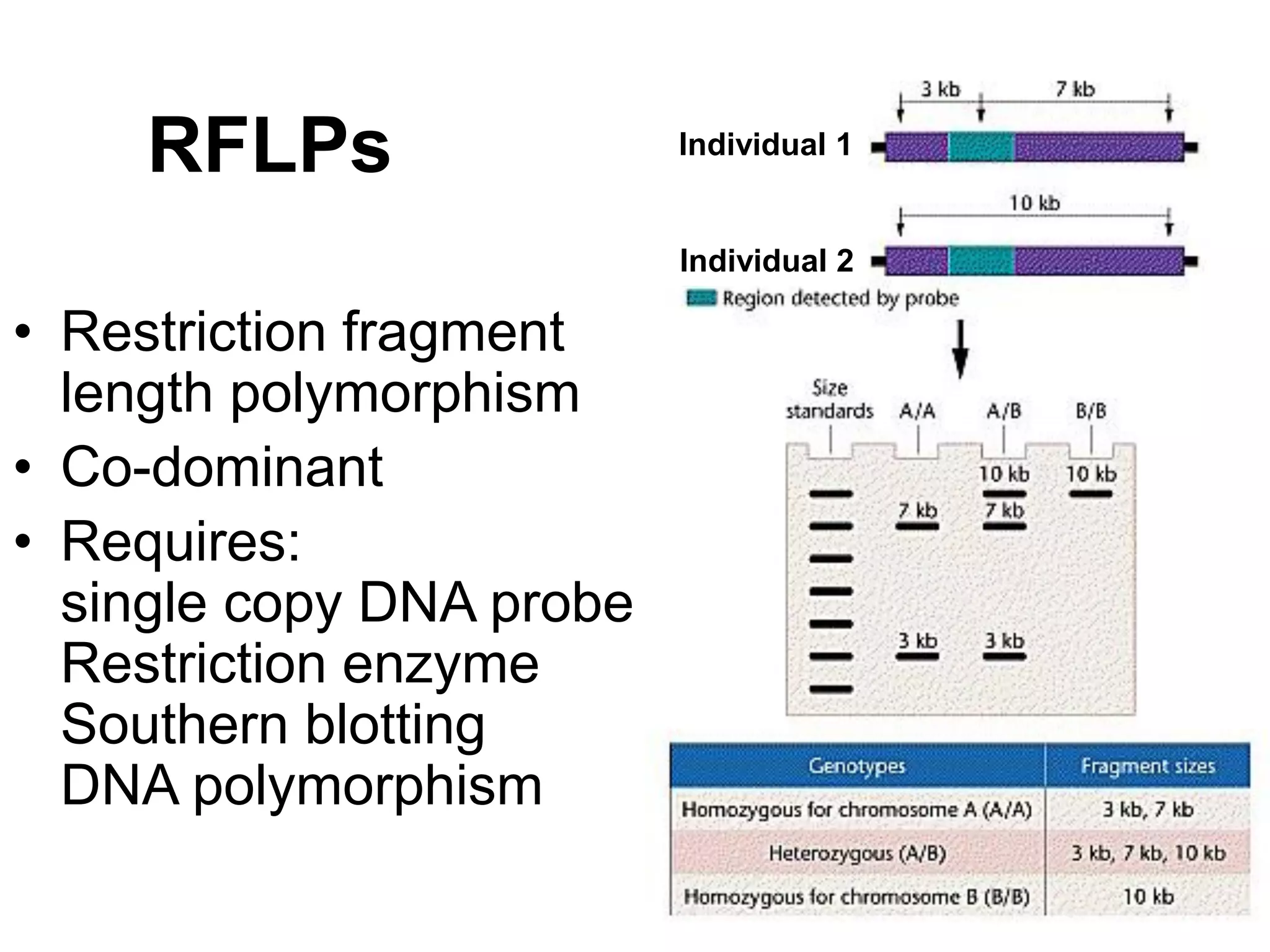
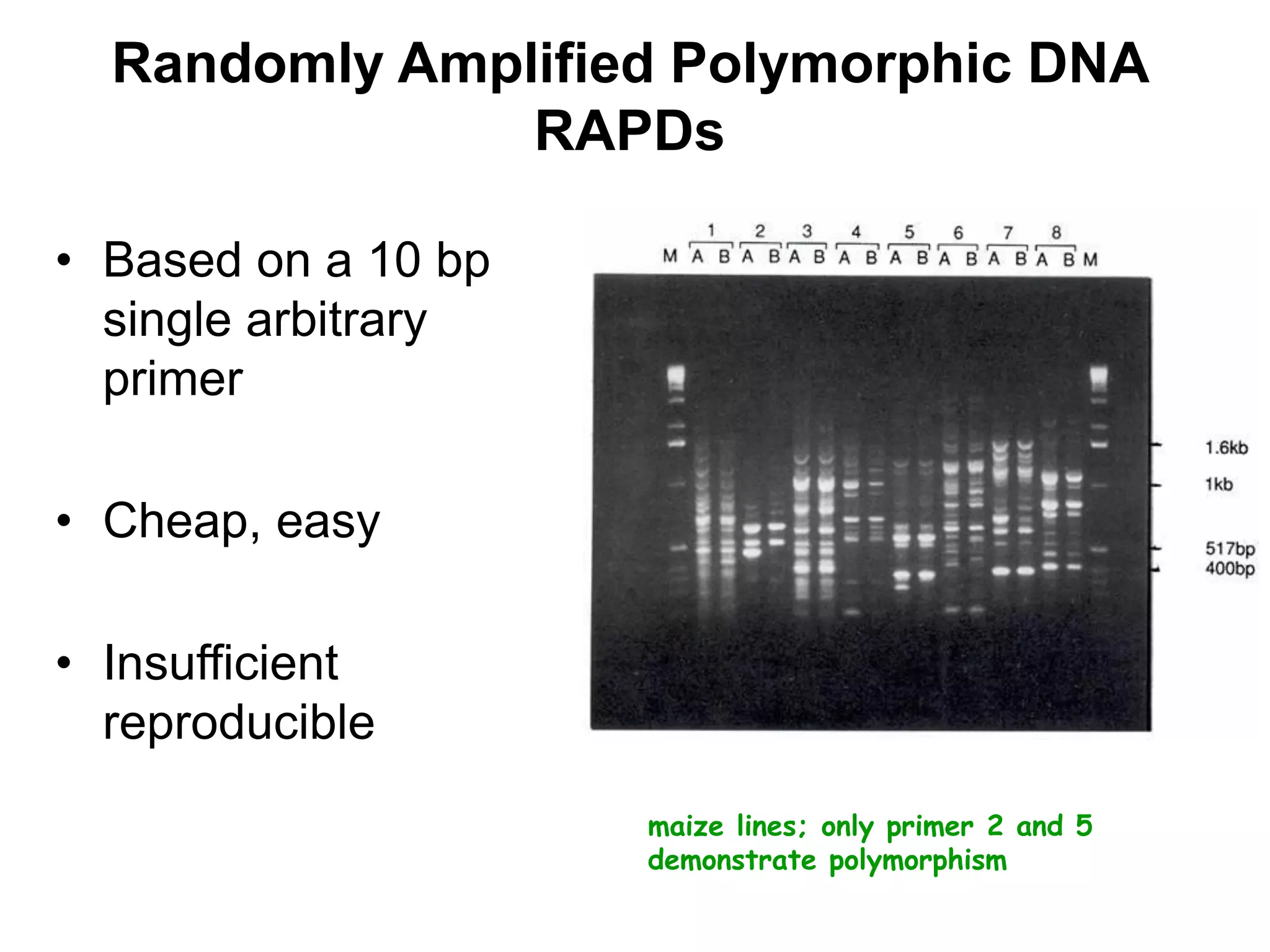
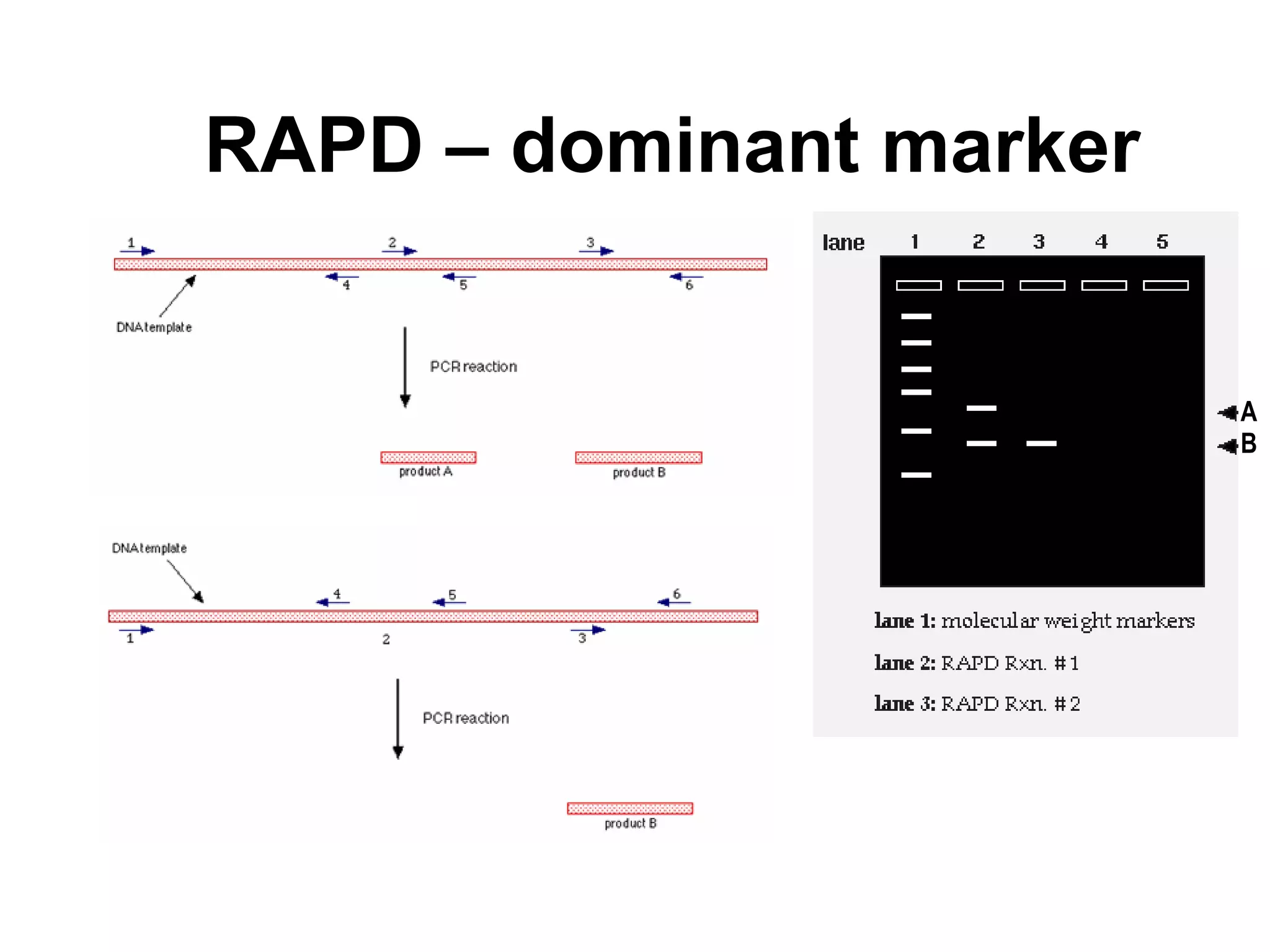
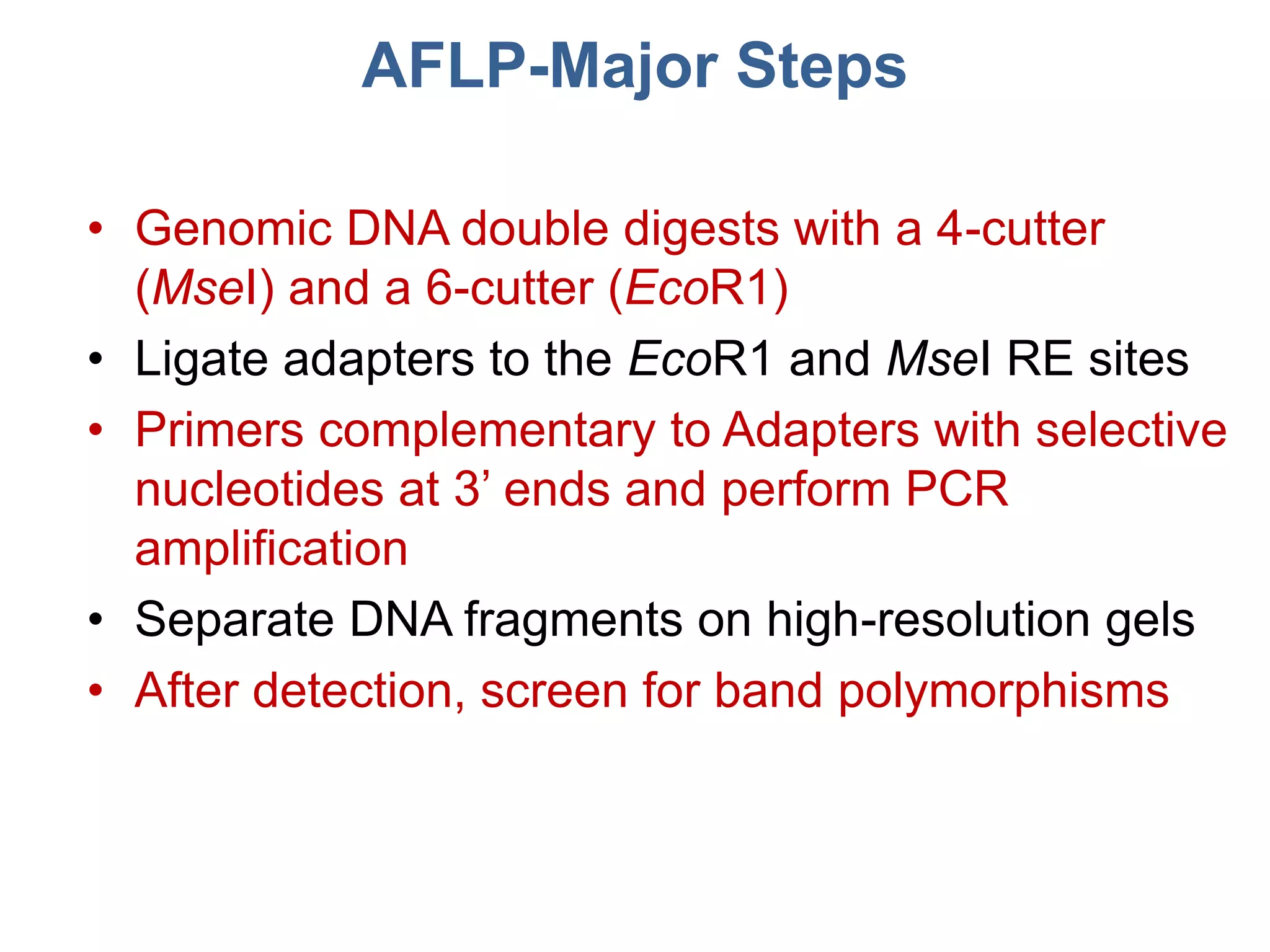
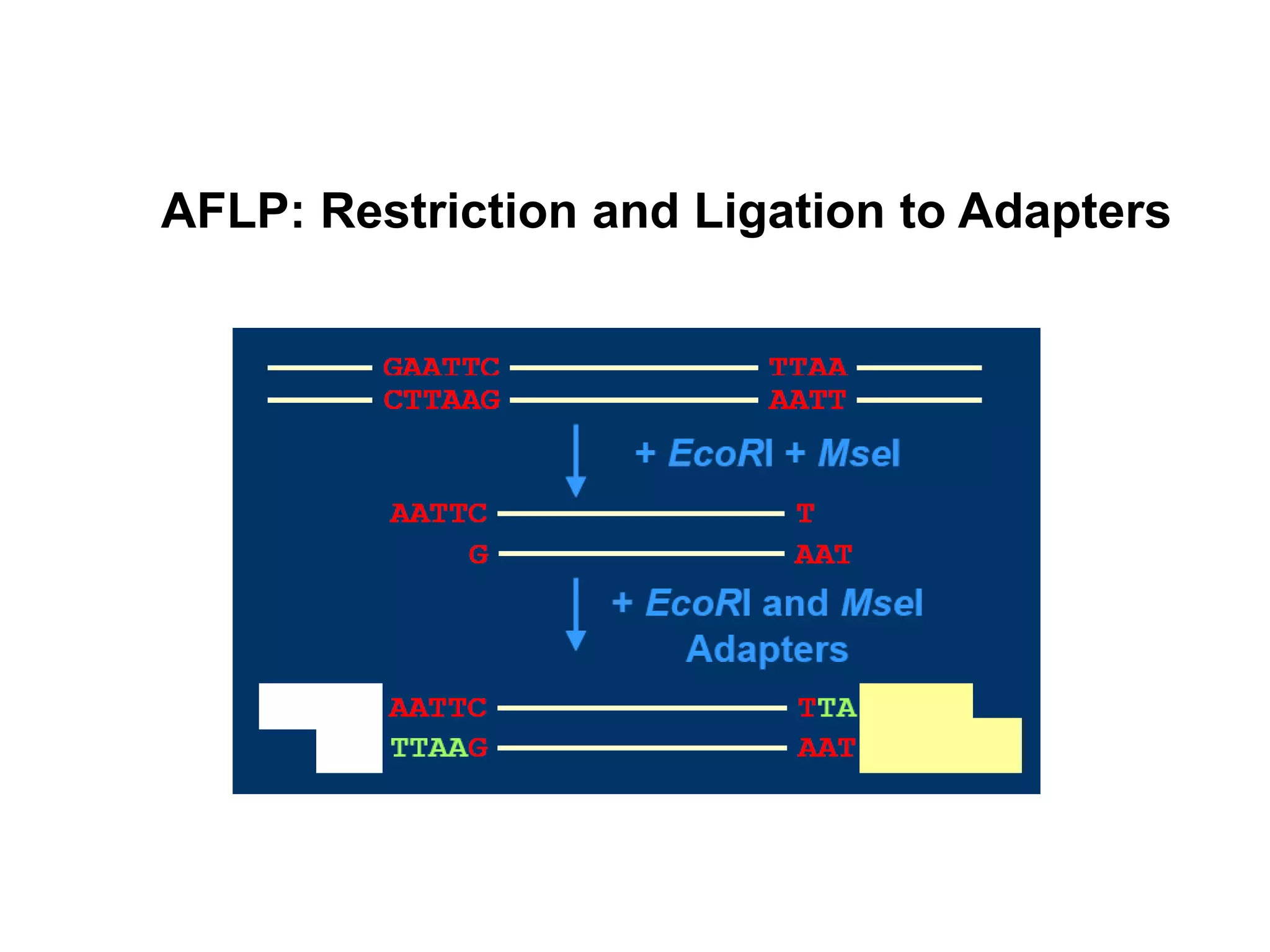
This document discusses molecular markers, which are short DNA sequences that reveal polymorphisms between individuals. Molecular markers include SNPs, insertions/deletions, and variations in simple sequence repeats. The document describes several types of molecular markers and methods used to detect them, including RFLPs, RAPDs, AFLPs, SSRs, ISSRs, and CAPS. The applications of molecular markers include paternity testing, disease diagnosis, forensic analysis, and varietal identification in plants.

![SSR - methodolgy
Genotype A
Genotype A Genotype B
PCR amplification with
radiolabelled nucleoltide
Polyacrylamide Gel Electrophoresis
of PCR products and autoradiography
[AT]18 [AT]22
[AT]
18
[AT]
22
Genotype B](https://image.slidesharecdn.com/molecularmarkersandapplications-lecture-230808071050-73ce0f3c/75/Molecular-Markers-and-Applications-Lecture-pdf-24-2048.jpg)