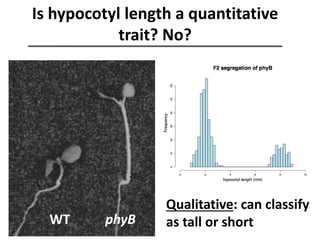
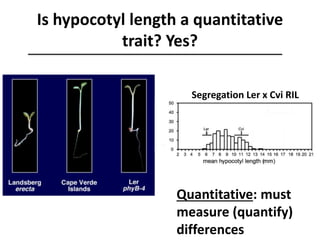
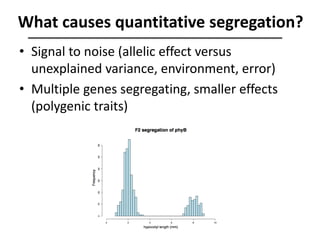
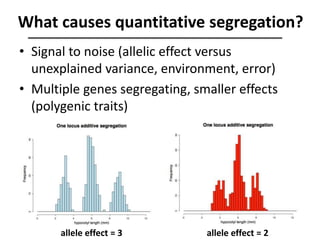
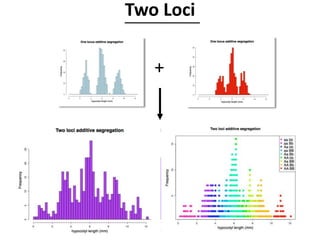
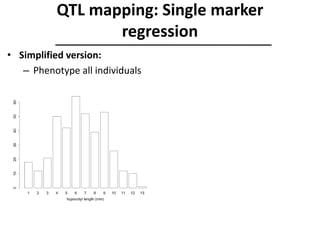
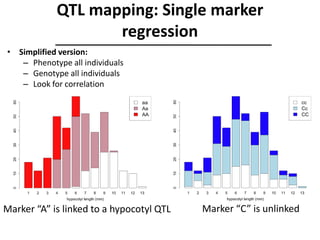
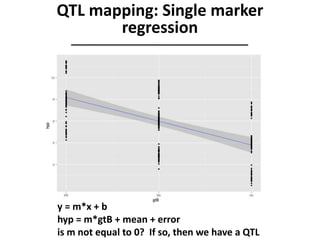
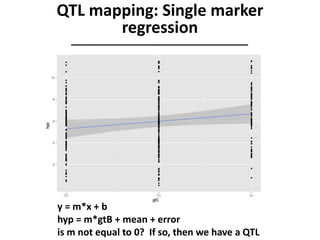
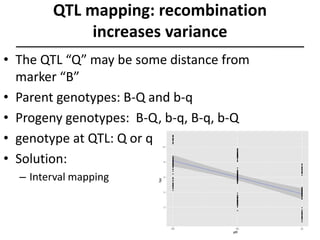
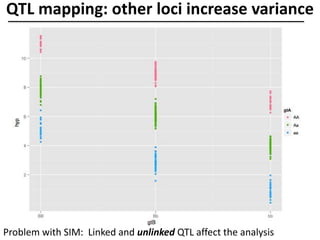
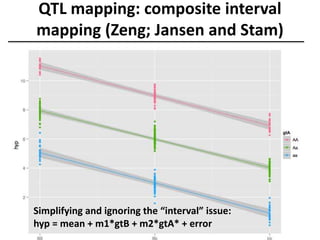
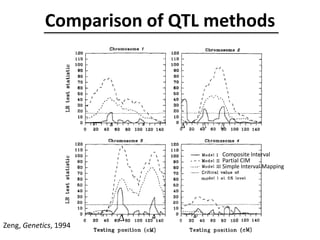
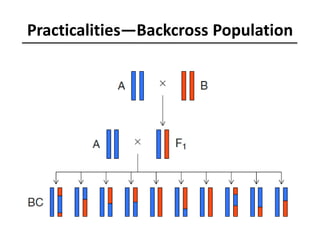
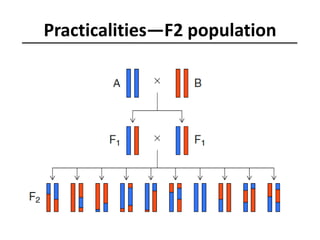
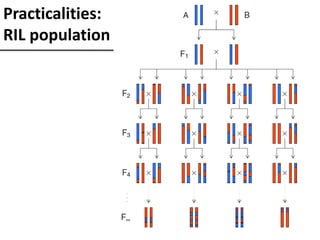
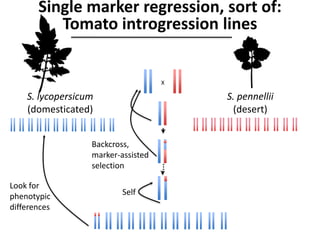
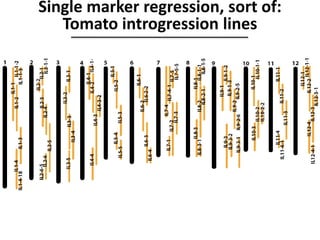
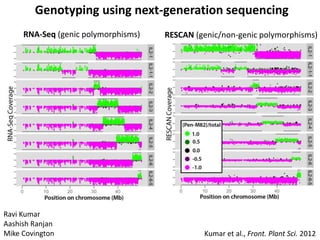
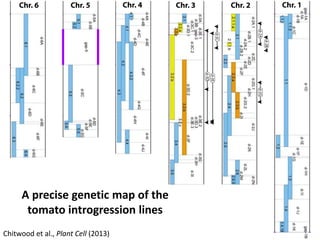
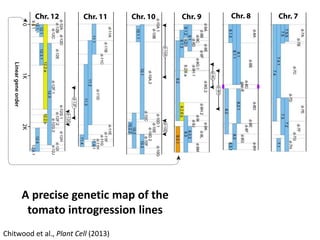
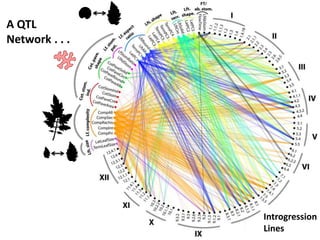
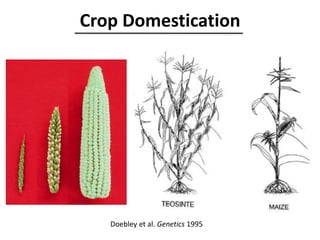
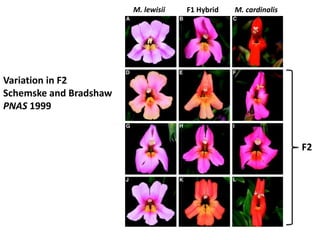
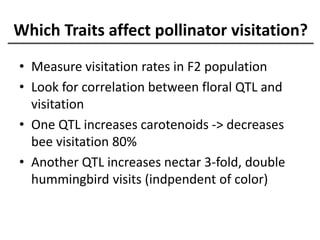
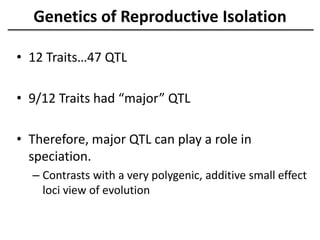
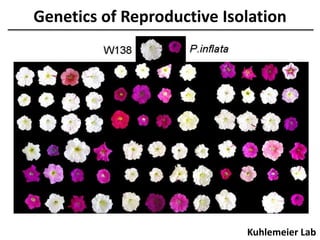
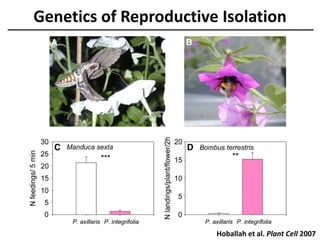
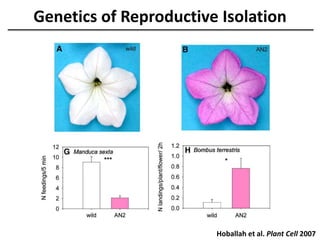
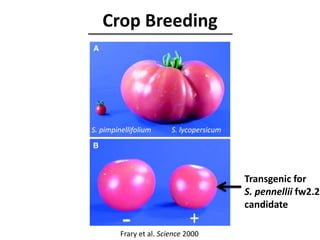
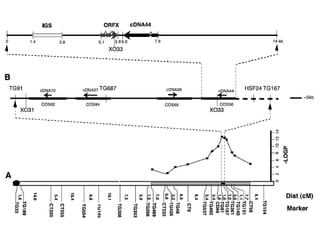
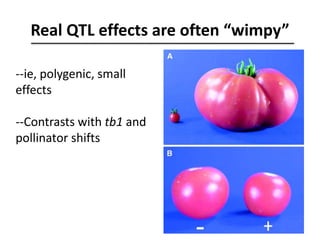
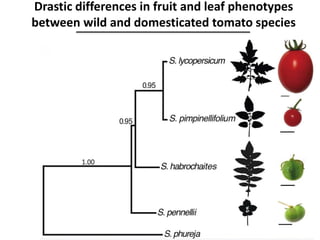
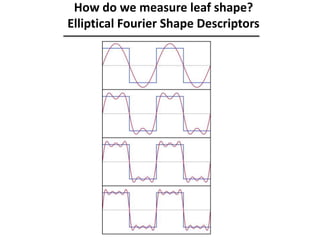
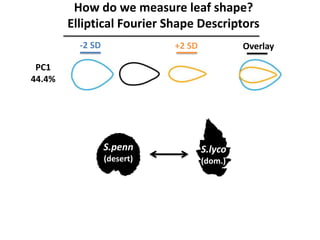
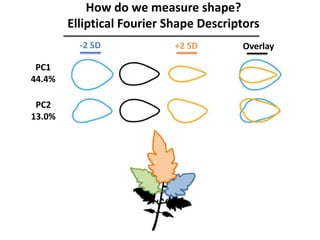
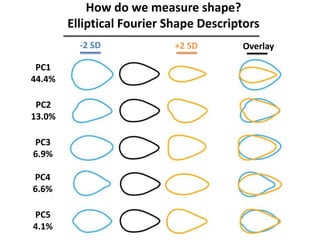
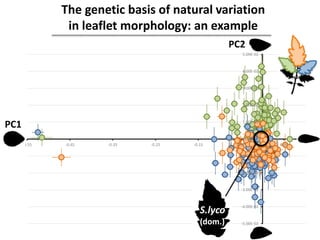
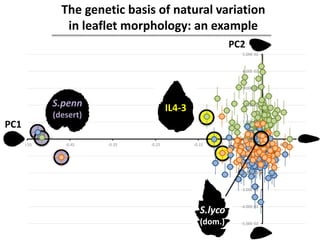
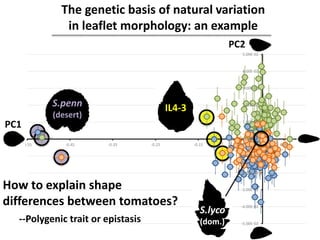
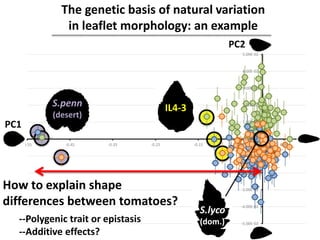
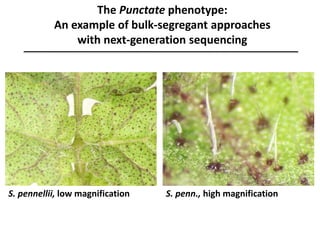
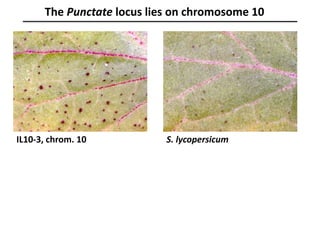
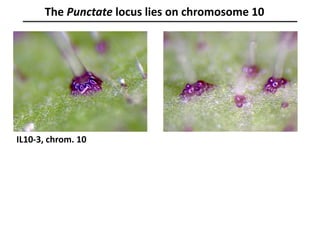
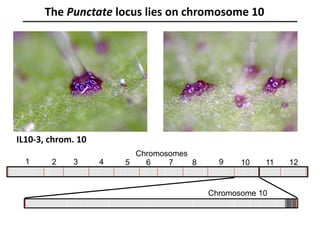
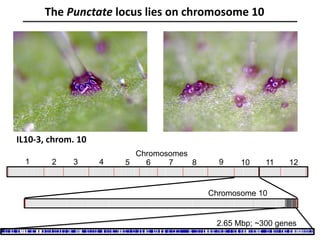
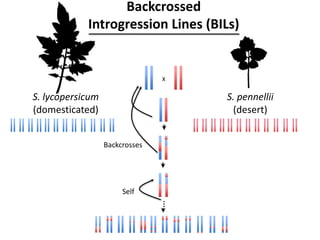
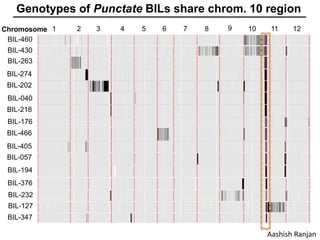
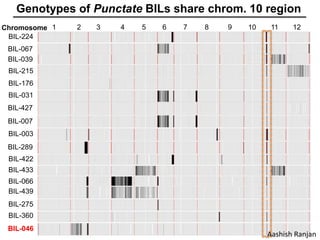
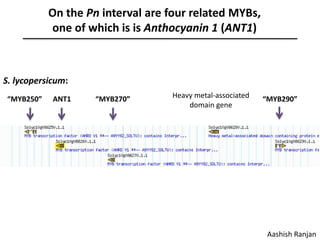
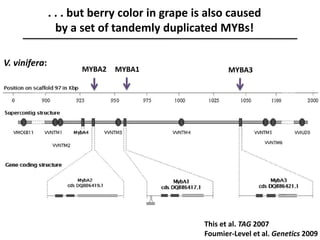
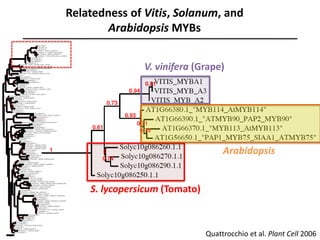
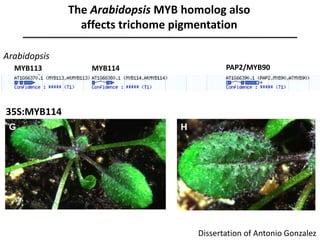
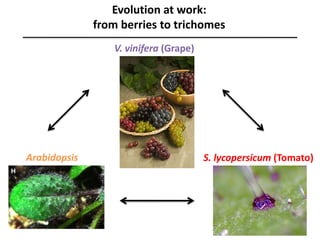
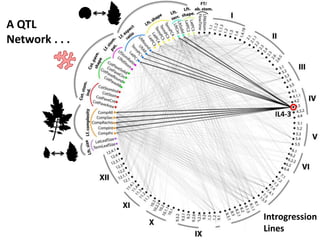
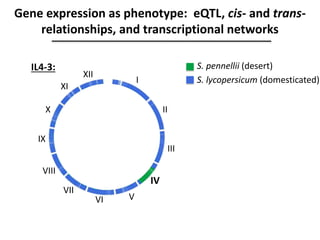
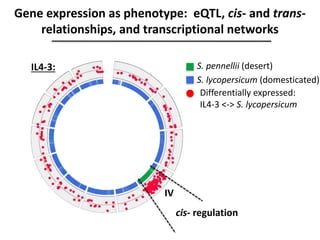
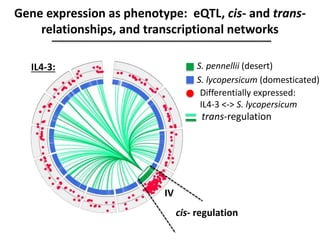
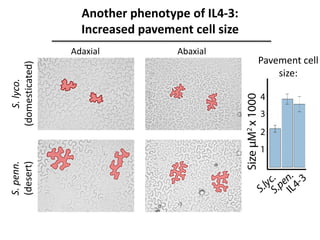
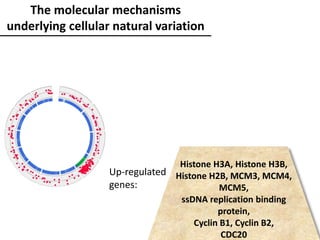
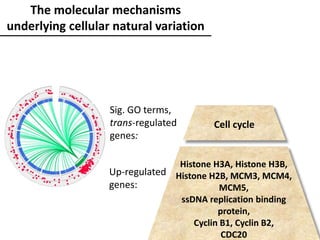
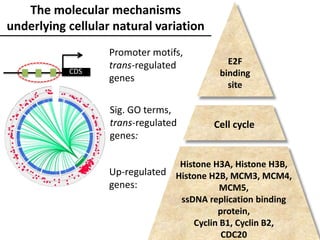
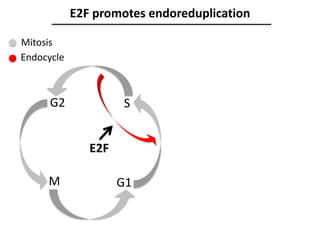
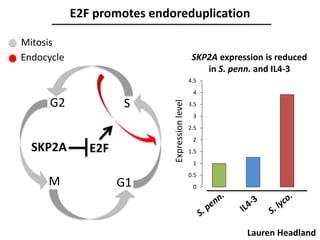
The document provides an introduction to quantitative genetics, focusing on quantitative trait loci (QTLs) and their role in traits like hypocotyl length. It discusses various methods for QTL mapping, such as single marker regression and interval mapping, while addressing challenges such as confounding factors and the effects of polygenic traits. Case studies in plant breeding, micro-evolution, and genetics of reproductive isolation are presented to illustrate the applications of QTL analysis.