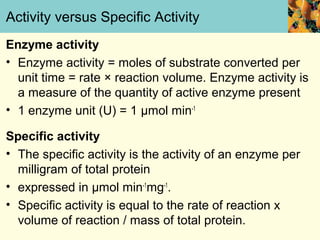
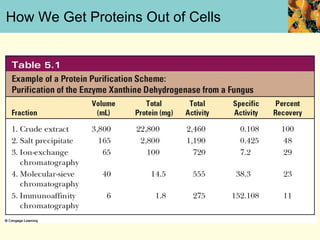
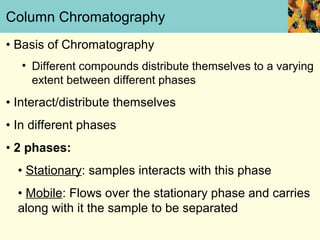
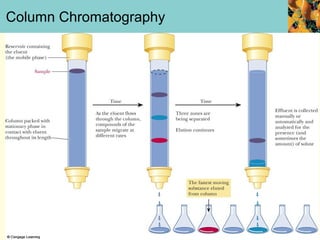
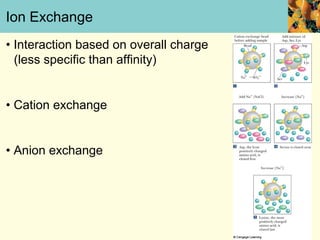
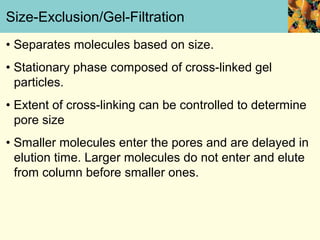
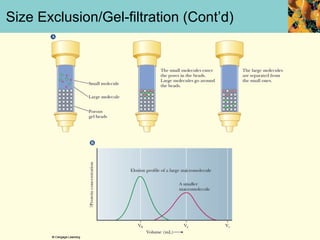
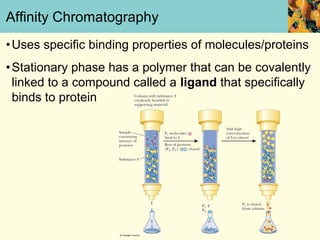
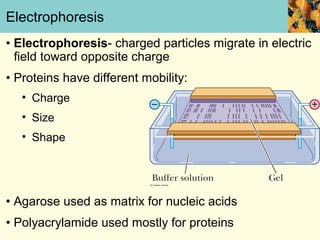
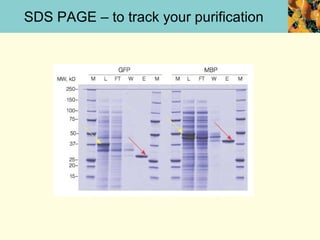
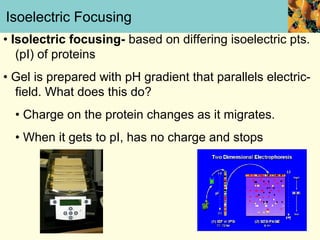
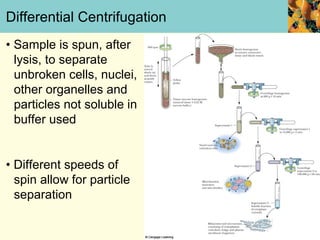
This document provides an overview of protein purification and characterization techniques, outlining the importance of defining objectives and properties of target proteins. It details various methods such as salting out, column chromatography, and electrophoresis, emphasizing the delicate nature of proteins and the need to track their activity during purification. Key metrics discussed include activity, specific activity, and the required purity for different applications.