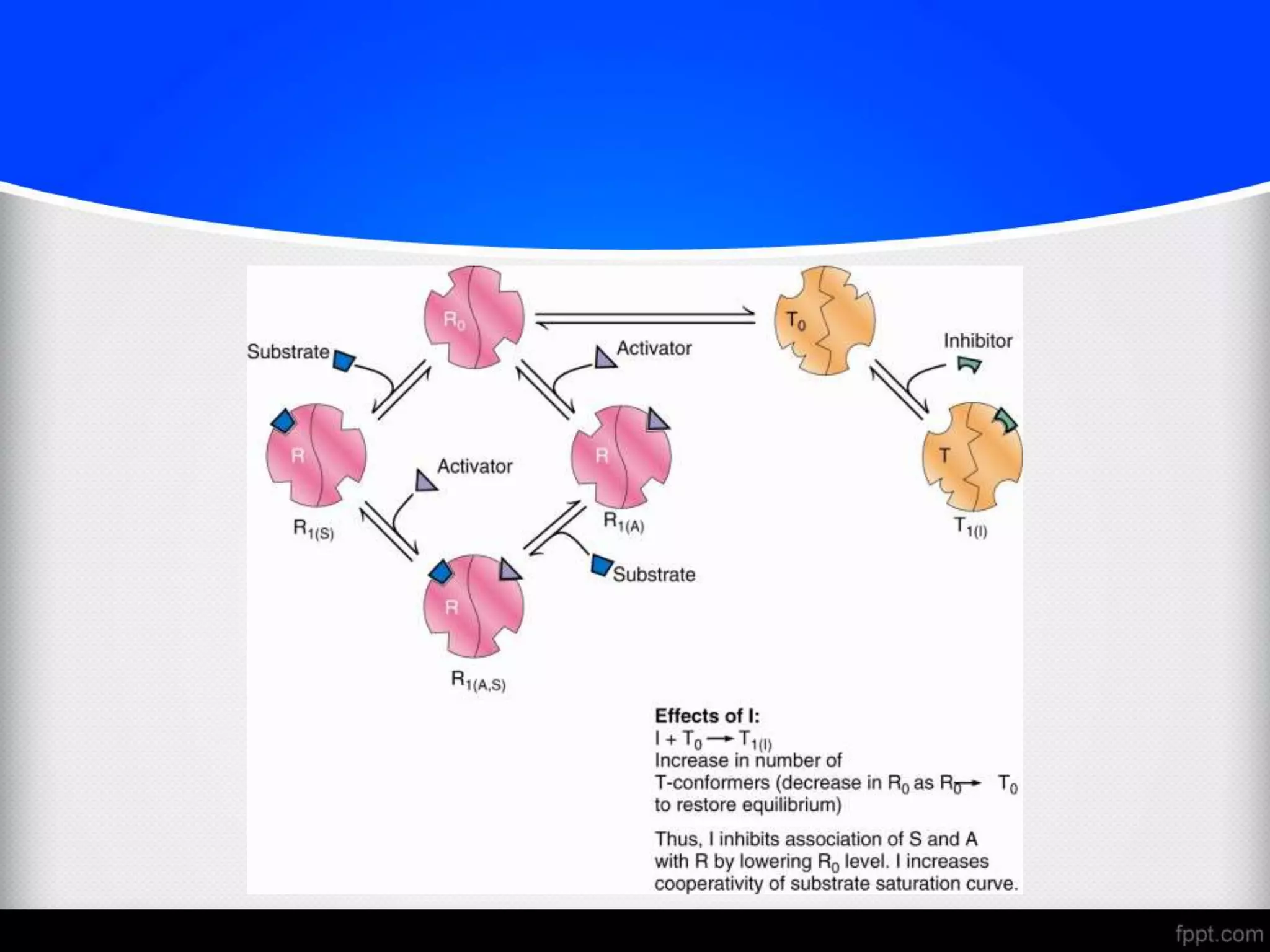
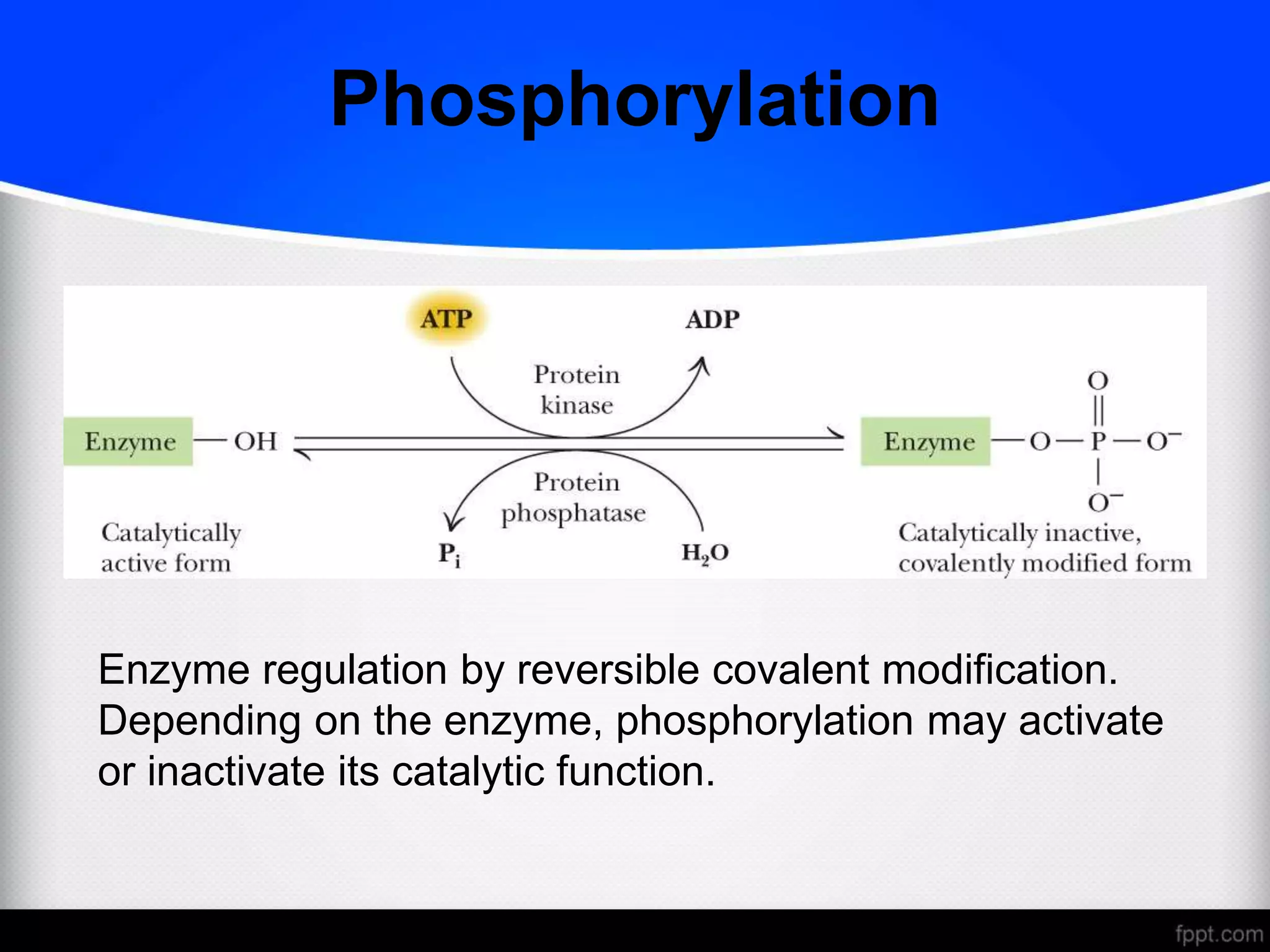
The document discusses various modes of enzyme regulation, including allosteric control, covalent modification, and hormonal influence, outlining their mechanisms and speeds of action. It details the roles of allosteric modulators, positive and negative effects on enzyme activity, and two models explaining allosteric interactions: the MWC concerted model and the KNF sequential model. Additionally, it covers genetic regulation of enzyme levels, highlighting constitutive and inducible enzymes, and the impact of protein processing on enzyme functionality.