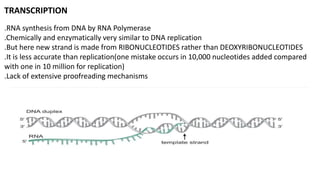
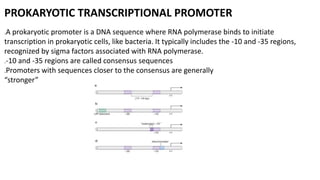
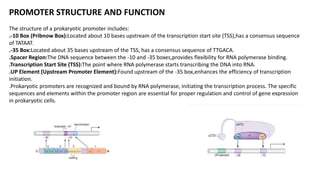
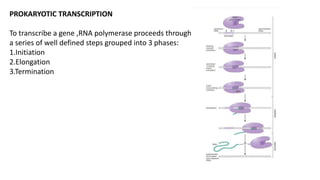
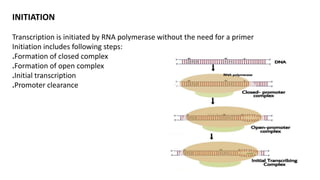
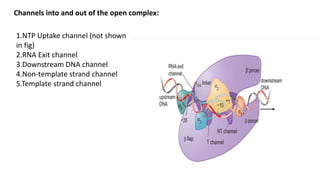
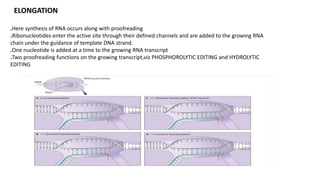
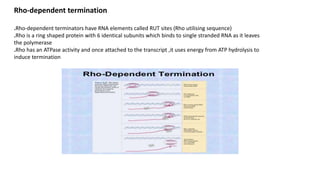
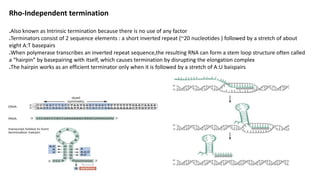
The document discusses prokaryotic transcription, focusing on the structure and function of promoters that RNA polymerase recognizes to initiate transcription in bacteria. It explains the key elements of promoters, such as the -10 and -35 consensus sequences, their roles in gene expression regulation, and the transcription process phases: initiation, elongation, and termination. Various termination mechanisms, including rho-dependent and rho-independent, are also summarized.