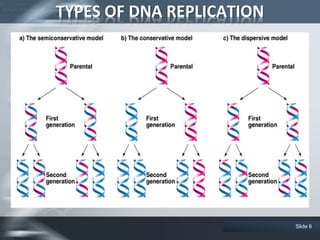
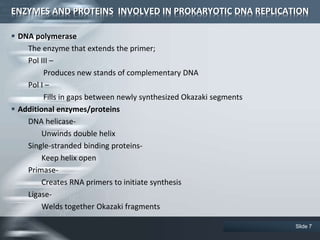
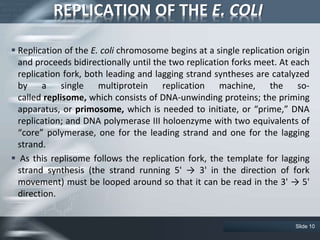
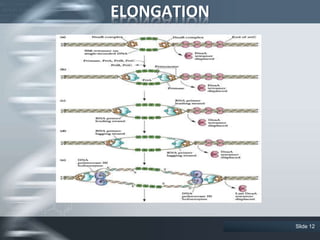
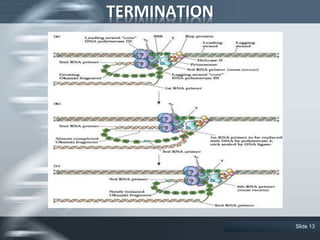
The document provides an overview of DNA replication, including its definition, historical milestones, and the types of enzymes and proteins involved in prokaryotic DNA replication. Key processes such as initiation, elongation, and termination are discussed, along with the structure of the replication fork and the role of the replisome during the replication of E. coli chromosomes. The conclusion emphasizes the possibility of in vitro DNA replication techniques, particularly the polymerase chain reaction (PCR).