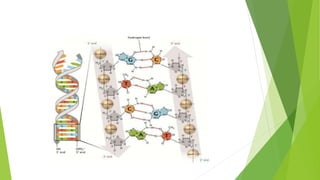
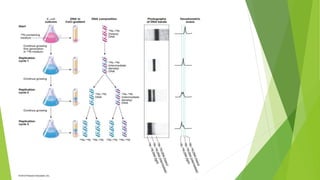
The document provides a comprehensive overview of DNA replication, detailing the Watson-Crick model, various replication modes (conservative, semi-conservative, dispersive), and the Meselson-Stahl experiment that supports semi-conservative replication. It lists key enzymes involved in the replication process, such as helicase and DNA polymerases, and outlines the steps of replication, including replication fork formation, elongation, and termination. Additionally, it highlights the differences in DNA replication between prokaryotes and eukaryotes.