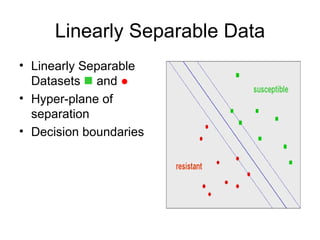
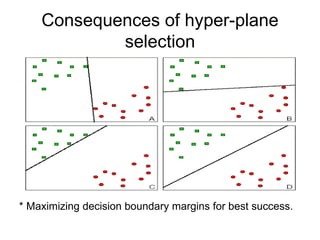
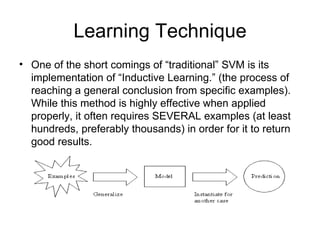
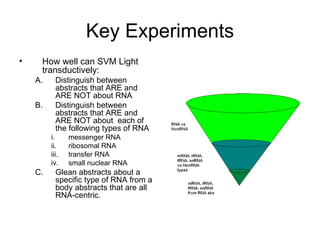
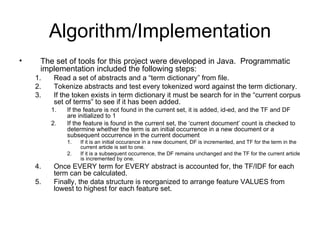
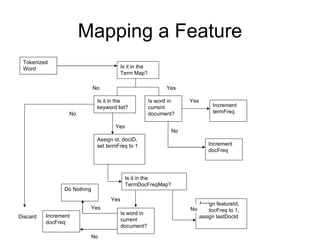
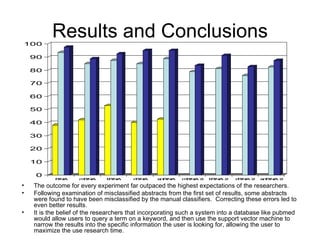
The document summarizes a graduate student's project using support vector machines (SVM) for transductive learning to classify RNA-related biological abstracts. The student collected a corpus of 400 abstracts categorized into RNA-related and non-RNA-related groups. Software was developed to preprocess the abstracts, extract features, generate training and test sets for SVM Light, and test its ability to classify abstracts into different RNA categories like mRNA, tRNA, etc. The goal was to improve on keyword searches by using a small number of training examples from a specific dataset to maximize classification precision for that set.