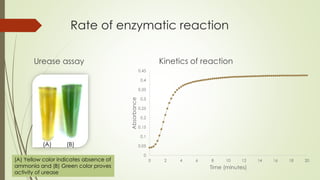
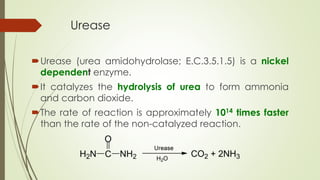
The document describes the purification of urease from sword bean for potential clinical use. Several issues were encountered with a previous purification method using an affinity matrix. The current study aimed to develop a simpler and lower cost purification process using solvent extraction followed by isoelectric focusing. Various parameters of the extraction process were optimized to address issues like inhibitor presence, activity loss, pH, and microbial growth. The purified urease was shown to retain activity and was stable for several weeks. It was used to prepare a low-cost urea test kit that demonstrated stability of results for over a month.



![Previous work from our laboratory
Commercially available urease: Jack bean (not grow well in
Bangladesh).
Results of our screening experiments: watermelon, yard long
bean, arahar dal, sword bean etc.
Saem et al. [1] from our laboratory has purified this sword
bean urease by preparing a urease selective affinity
chromatographic matrix.
Affinity matrix itself became unusable after several
purification processes, thus increased the purification cost of
urease.
[1] Saem et al., (January, 2012) Isolation and affinity purification of urease from sword bean, M. Sc. Thesis, ACCE, RU.](https://image.slidesharecdn.com/885c302a-dcf8-4d76-a922-519a9e971d16-150802204757-lva1-app6892/85/Purification-of-Urease-from-Sword-Bean-for-Clinical-Use-5-320.jpg)





![Choice of solvent
SBM + 32% acetone
Supernatant
Centrifugation
There is a significant body of works on the solvent
extraction of jack bean urease and almost all the works
are based on the method of Sumner [1].
In his work, urease was extracted with 32% (v/v) acetone
solution of water at 28℃ and we have made a similar
approach.
Designation R1 (ml) Crude Extract (µl) Urea (µl) R2 (ml) Abs
Blank 1 100 - 1 0.098
Sample 1 100 10 1 0.156
[1] Sumner, J. B., (1951) Urease, In: Sumner, J. B. and Myrback, K. (Eds.) The enzymes, Academic Press,
New York, 873-892.](https://image.slidesharecdn.com/885c302a-dcf8-4d76-a922-519a9e971d16-150802204757-lva1-app6892/85/Purification-of-Urease-from-Sword-Bean-for-Clinical-Use-11-320.jpg)
![Remedy from inhibitor and activity loss
SBM + 32% acetone
+
1 mM EDTA and 1 mM
2-mercaptoethanol
Supernatant
Centrifugation
It has been reported that many divalent metal ions
inhibit urease in the following decreasing order [1]:
𝐻𝑔2+
> 𝐶𝑢2+
> 𝑍𝑛2+
> 𝐶𝑑2+
> 𝑁𝑖2+
> 𝑃𝑏2+
> 𝐶𝑜2+
> 𝐹𝑒2+
It has also been reported that JBU contains cysteine
residue near catalytic site [2] and it should most
probably be maintained in reduced condition for its
catalytic activity.
Designation R1 (ml) Crude Extract (µl) Urea (µl) R2 (ml) Abs
Blank 1 100 - 1 0.074
Sample 1 100 10 1 0.378
[1] Rezaei Behbehani et al., (2012) Inhibitory effect of cobalt (II) ion on jack bean urease, Res J Chem Sci, 2
(7), 72-74.
[2] Follmer et al., (2004) Jack bean urease (EC 3.5.1.5) aggregation monitored by dynamic and static light
scattering, Biophysical chemistry, 111 (1), 79-87.](https://image.slidesharecdn.com/885c302a-dcf8-4d76-a922-519a9e971d16-150802204757-lva1-app6892/85/Purification-of-Urease-from-Sword-Bean-for-Clinical-Use-12-320.jpg)