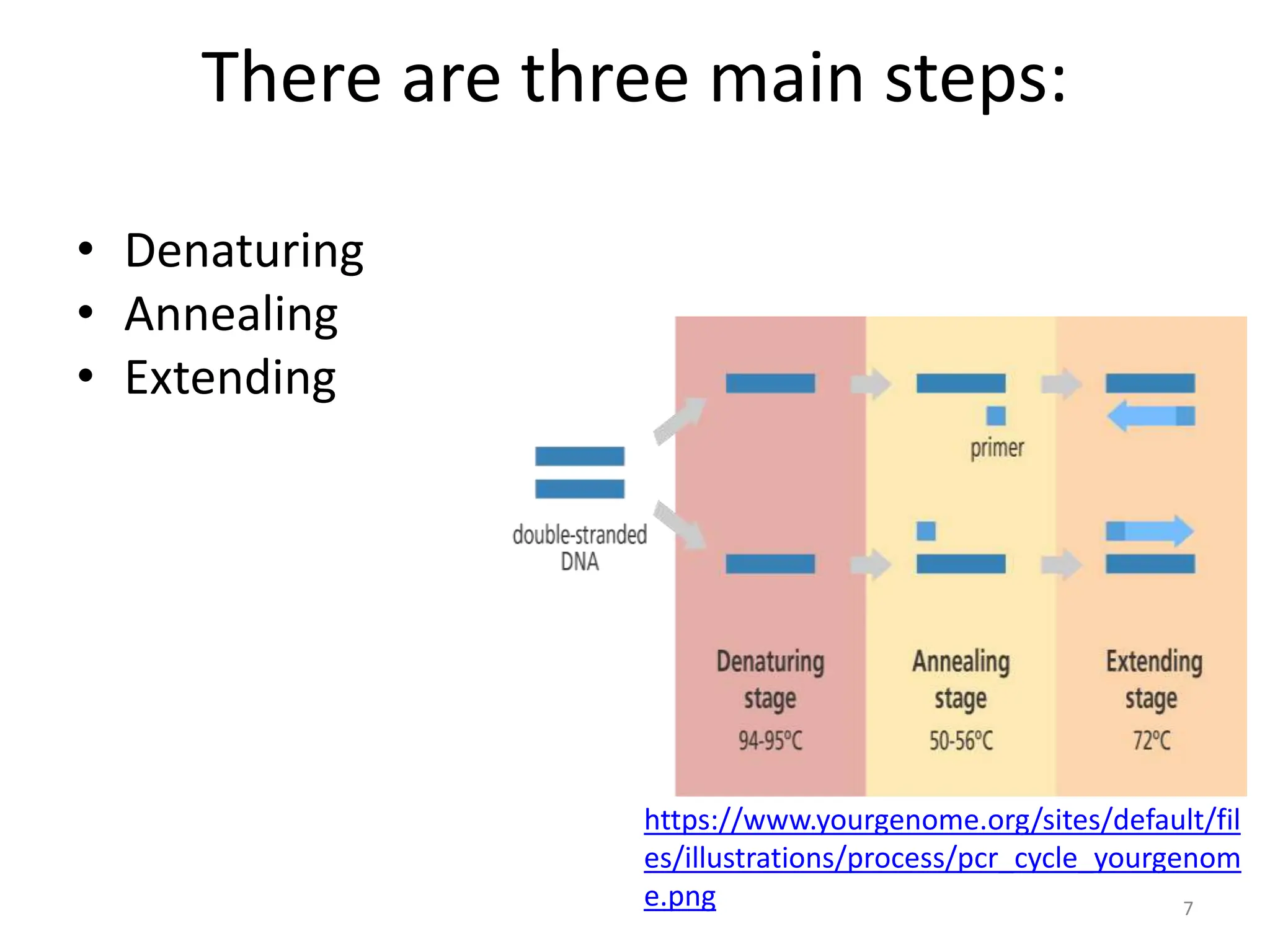
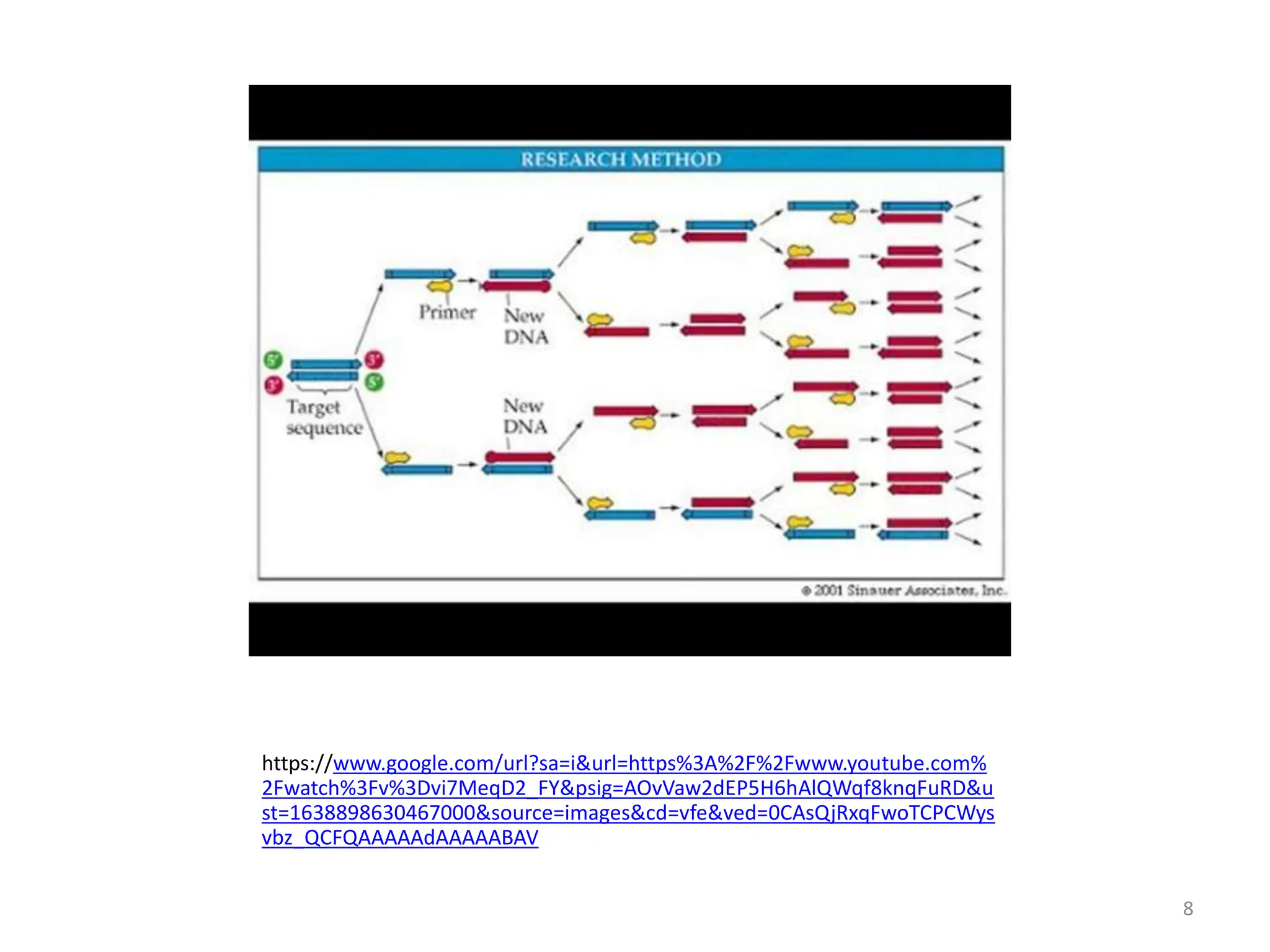
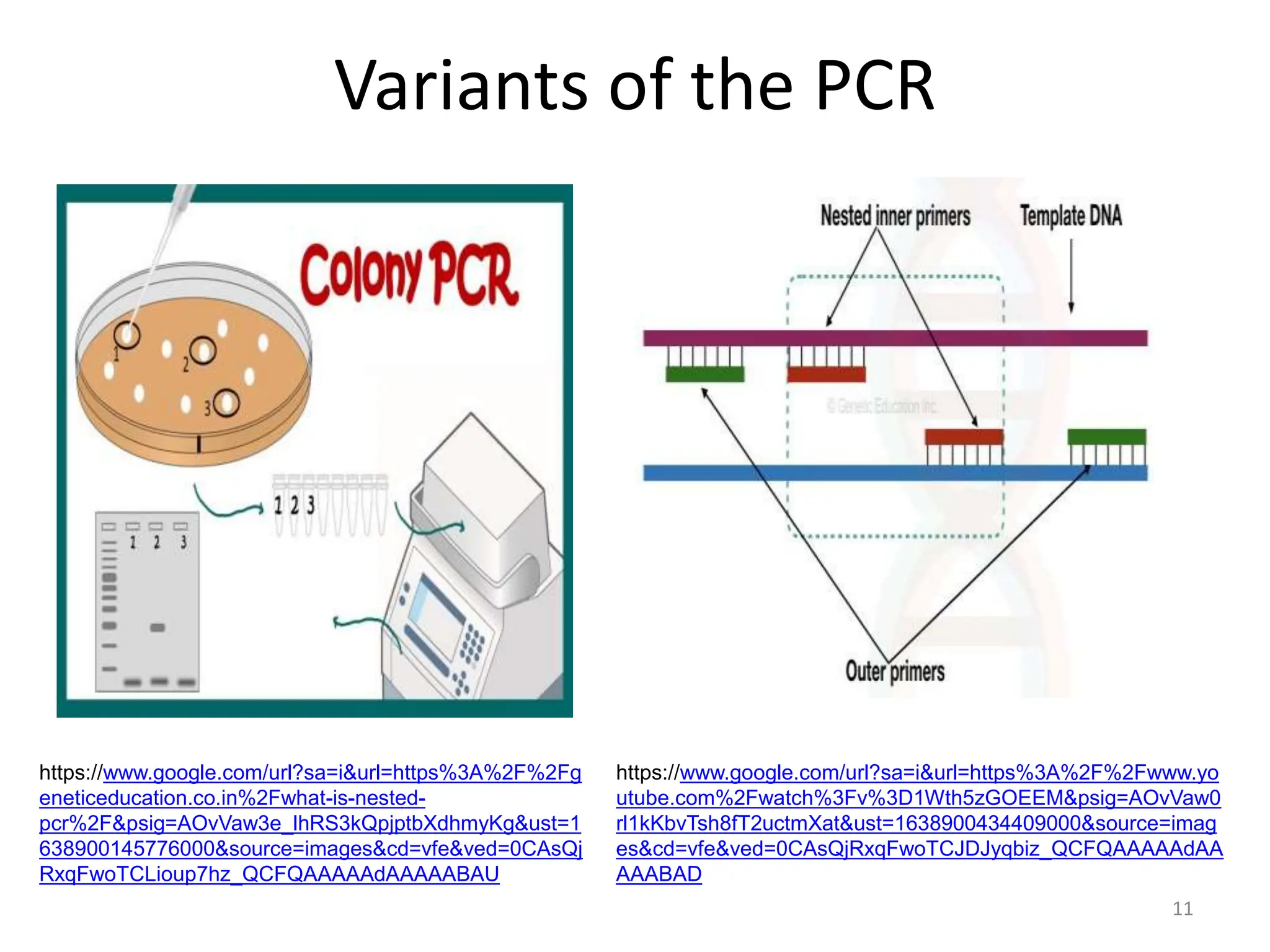
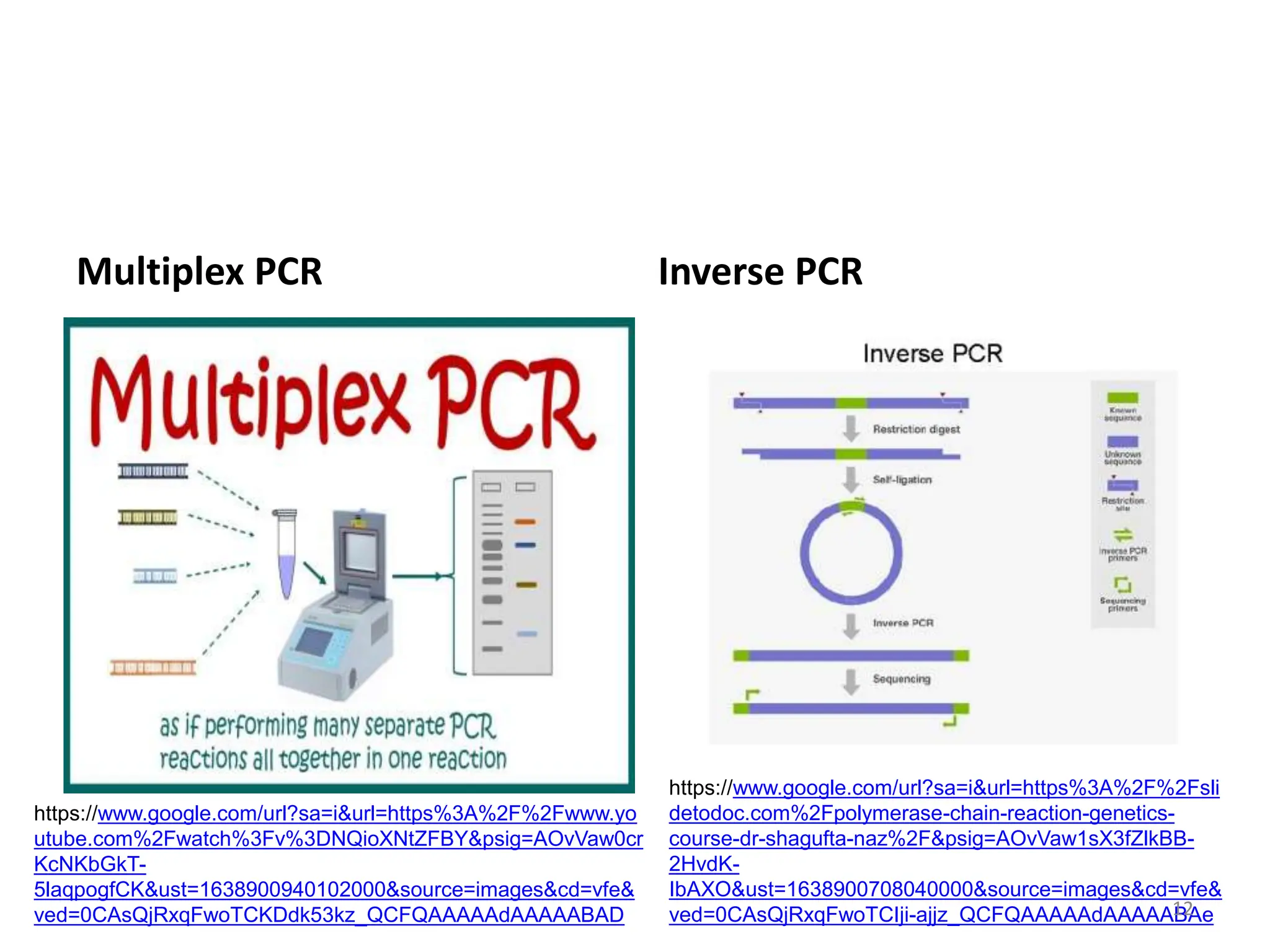
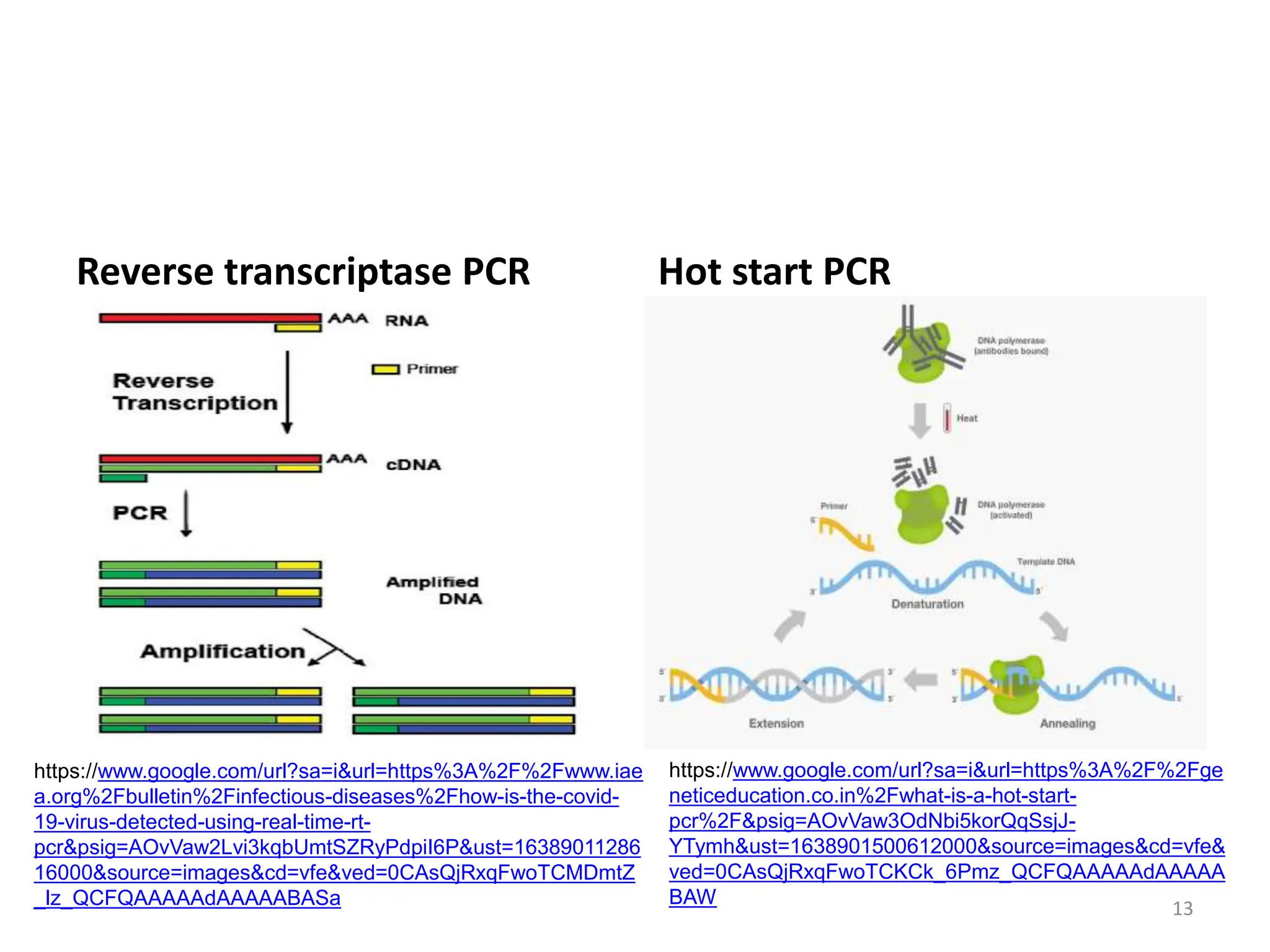
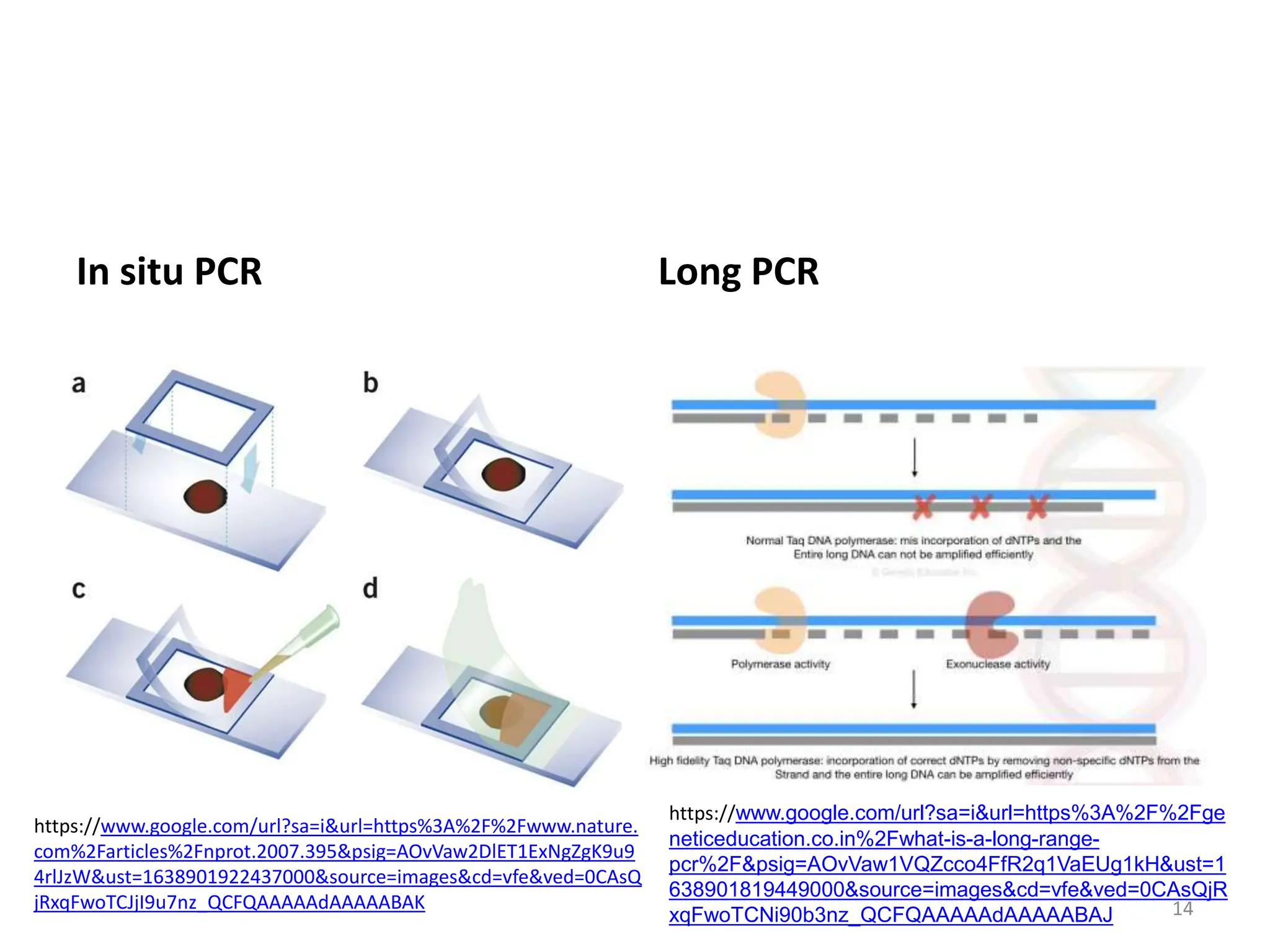
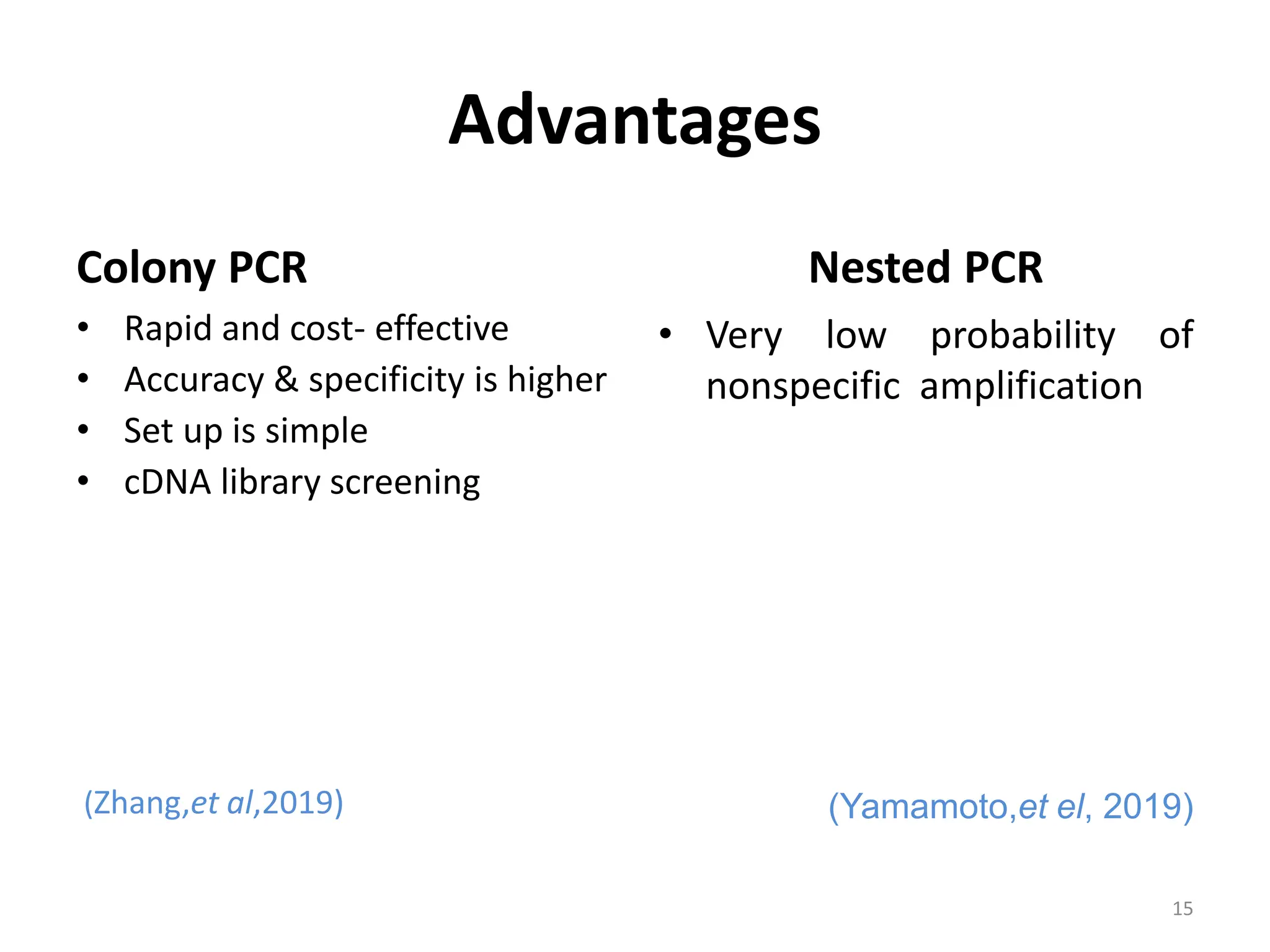
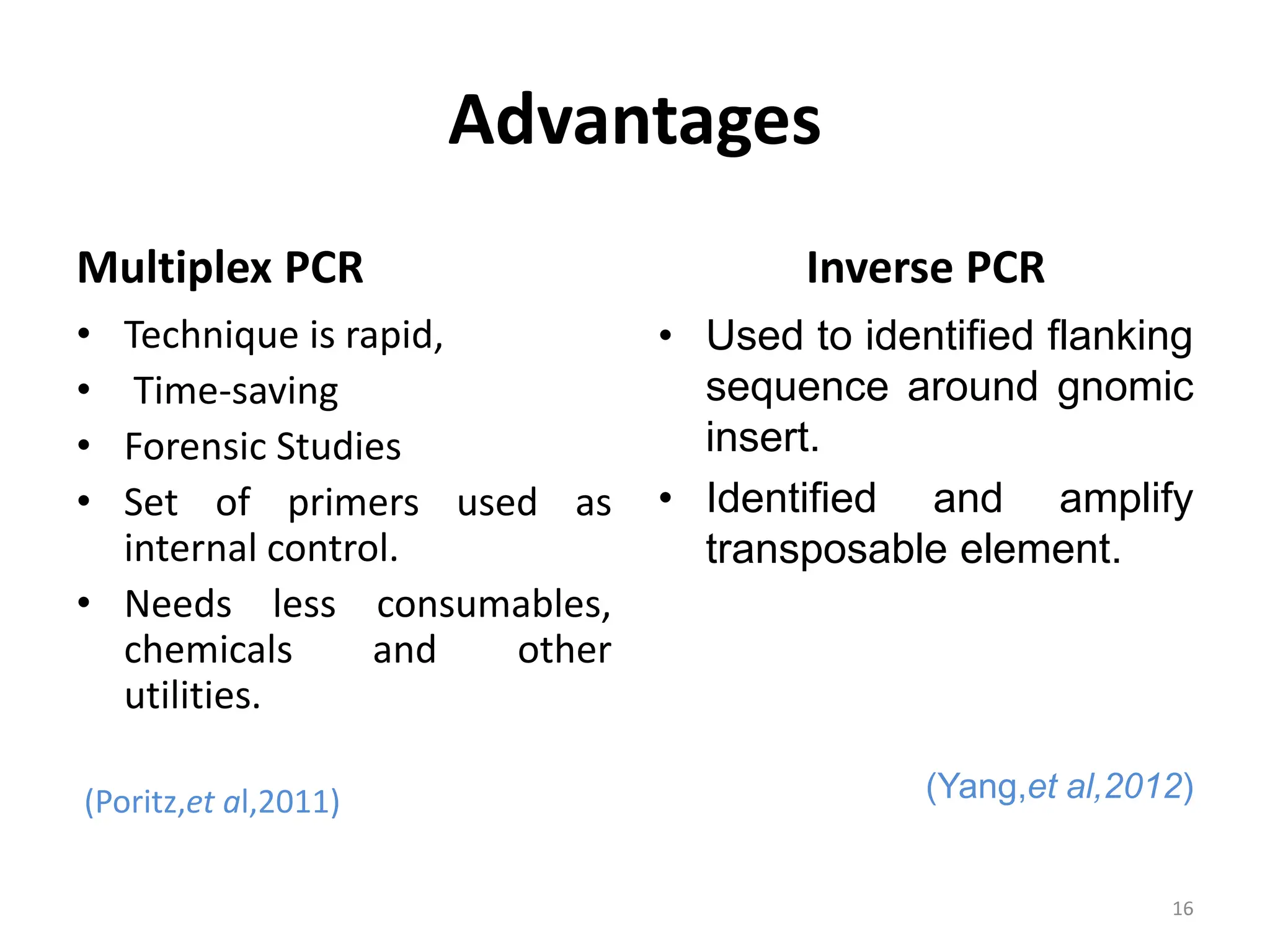
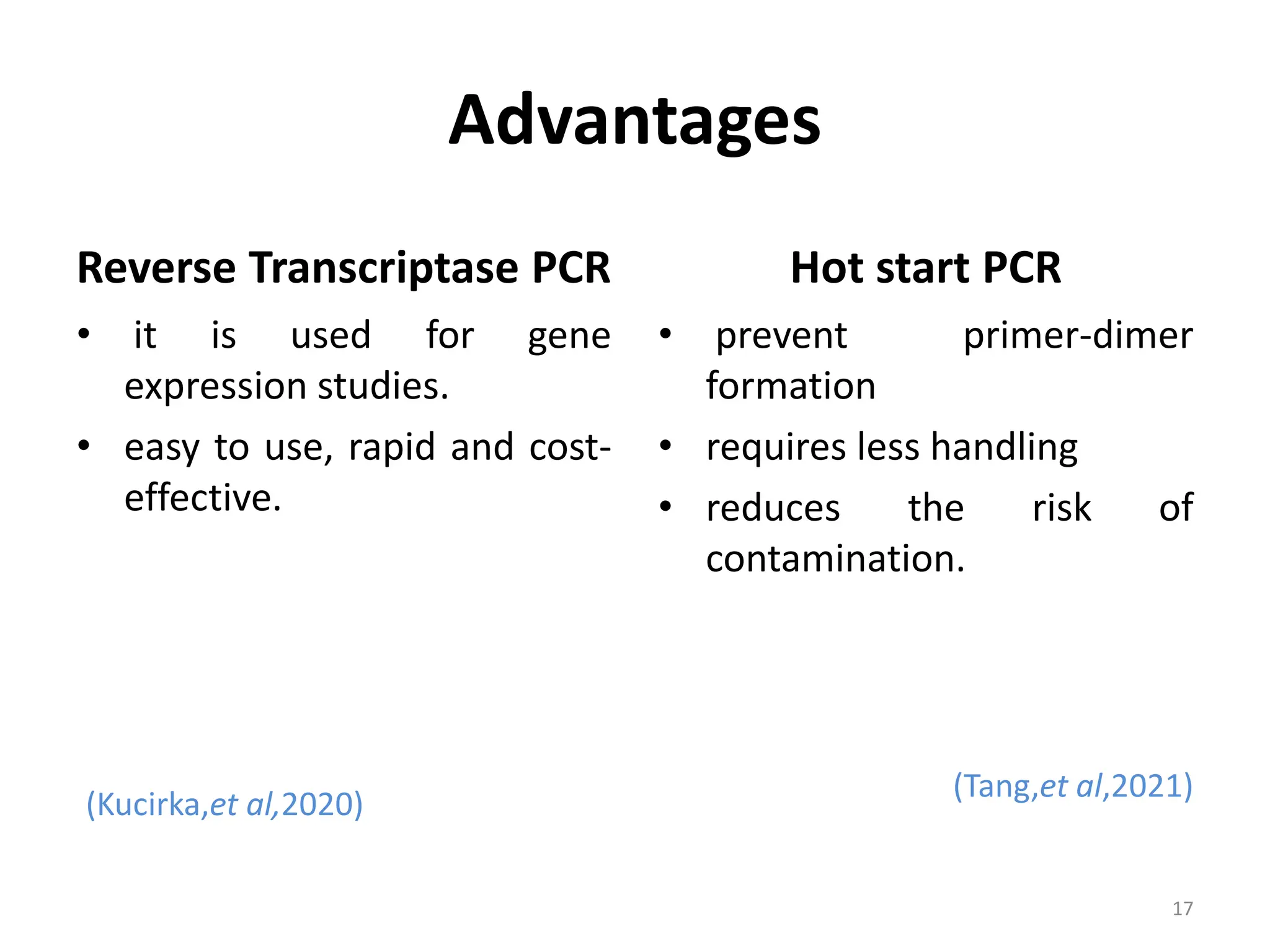
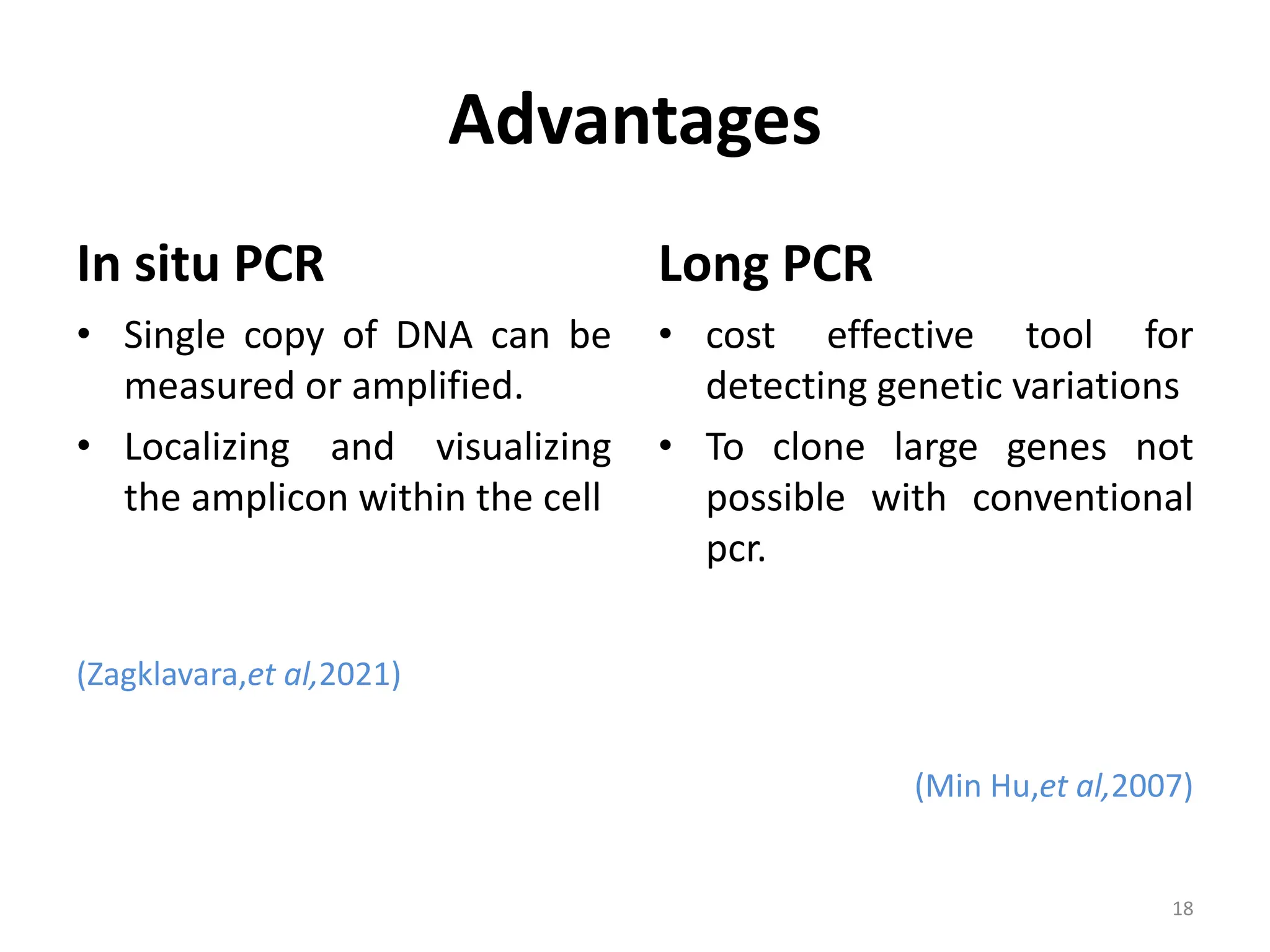
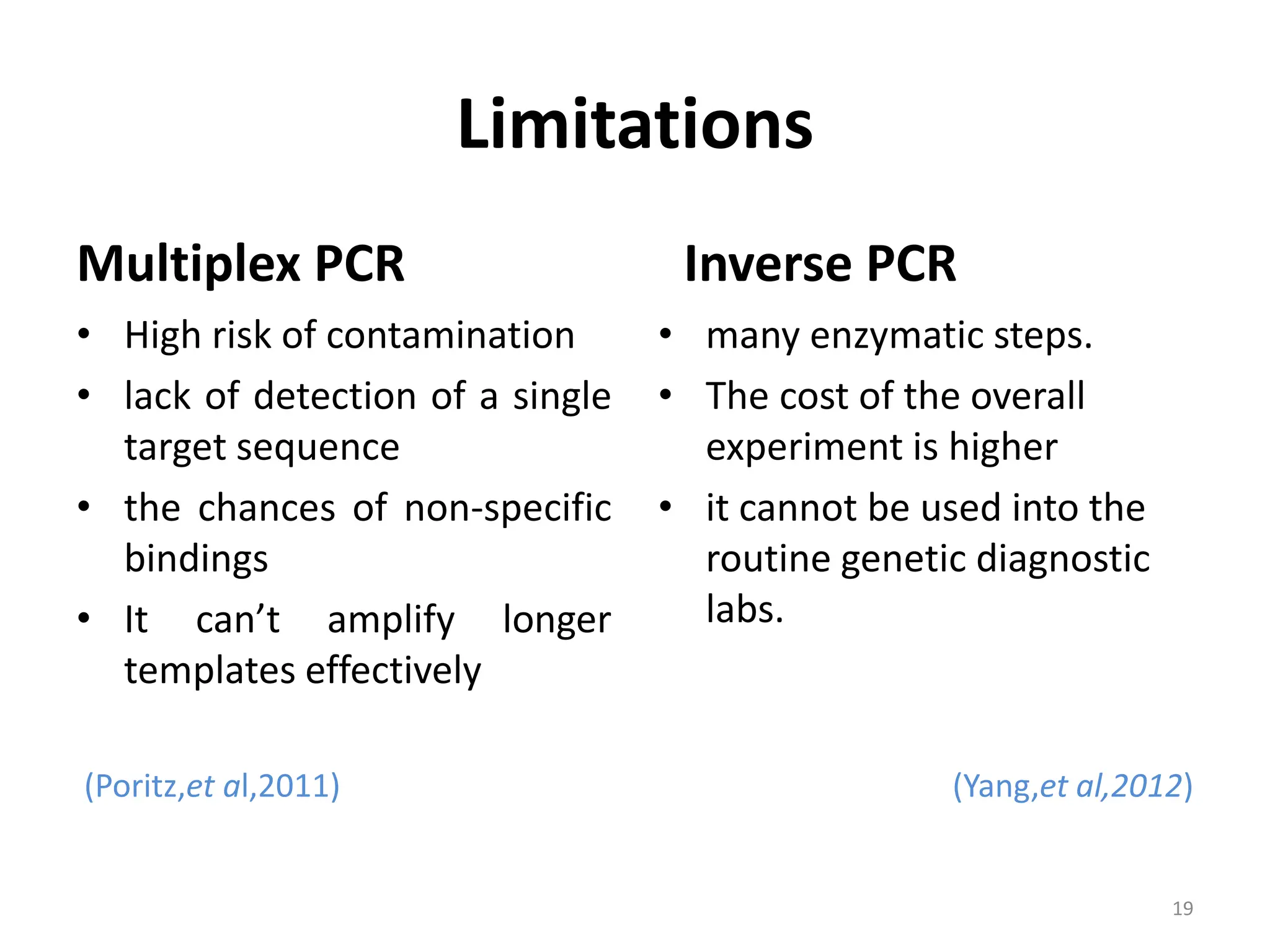
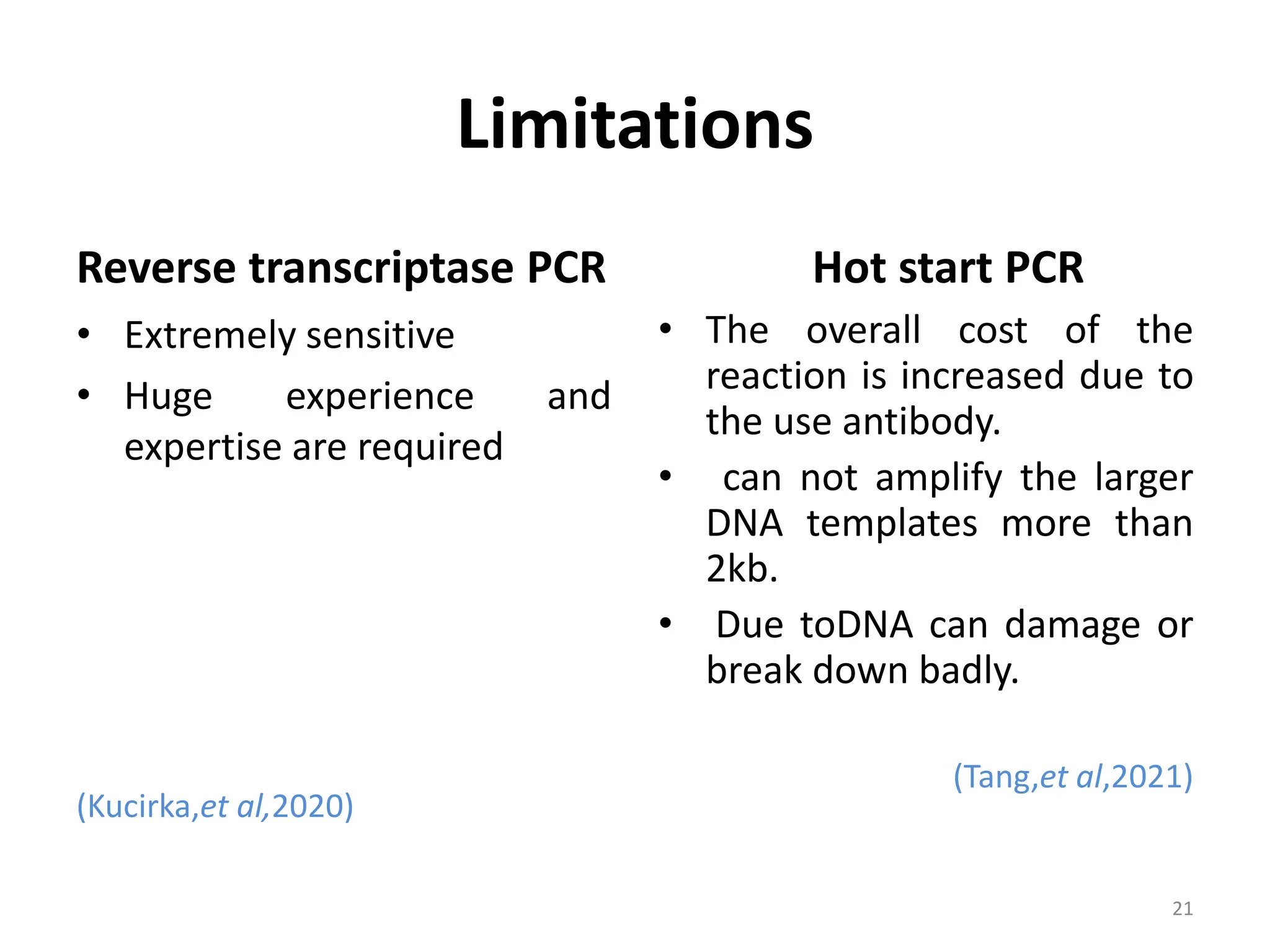
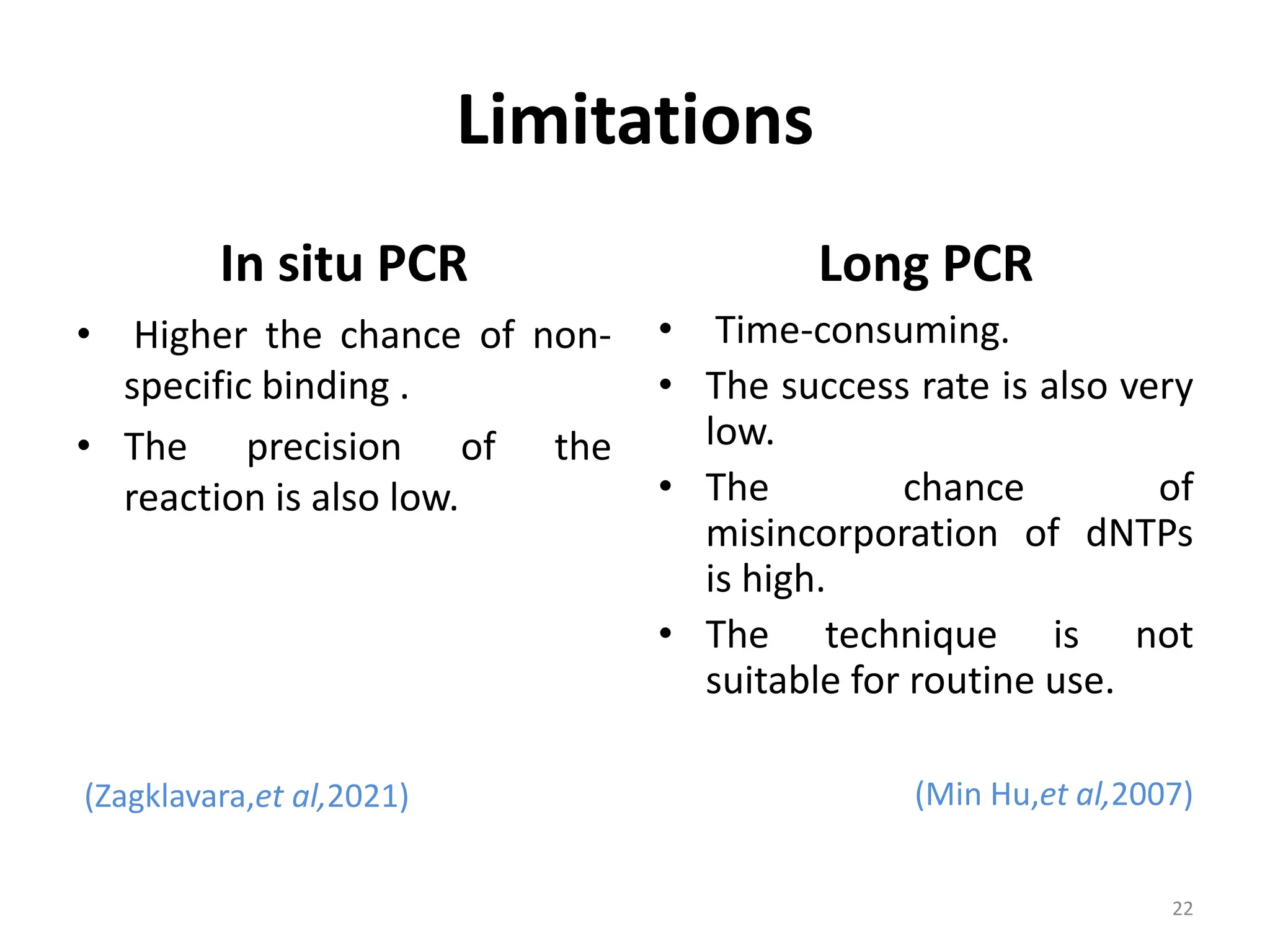
Polymerase Chain Reaction (PCR) is a technique used to make millions of copies of a specific DNA sequence. It was invented in 1983 by Kary Mullis. The basic steps of PCR include denaturation of DNA, annealing of primers, and extension of primers by DNA polymerase. There are many variants of PCR that have been developed, including real-time PCR, nested PCR, multiplex PCR, and long PCR. While PCR is a powerful technique, it also has limitations such as sensitivity to contamination and inability to amplify unknown DNA sequences.