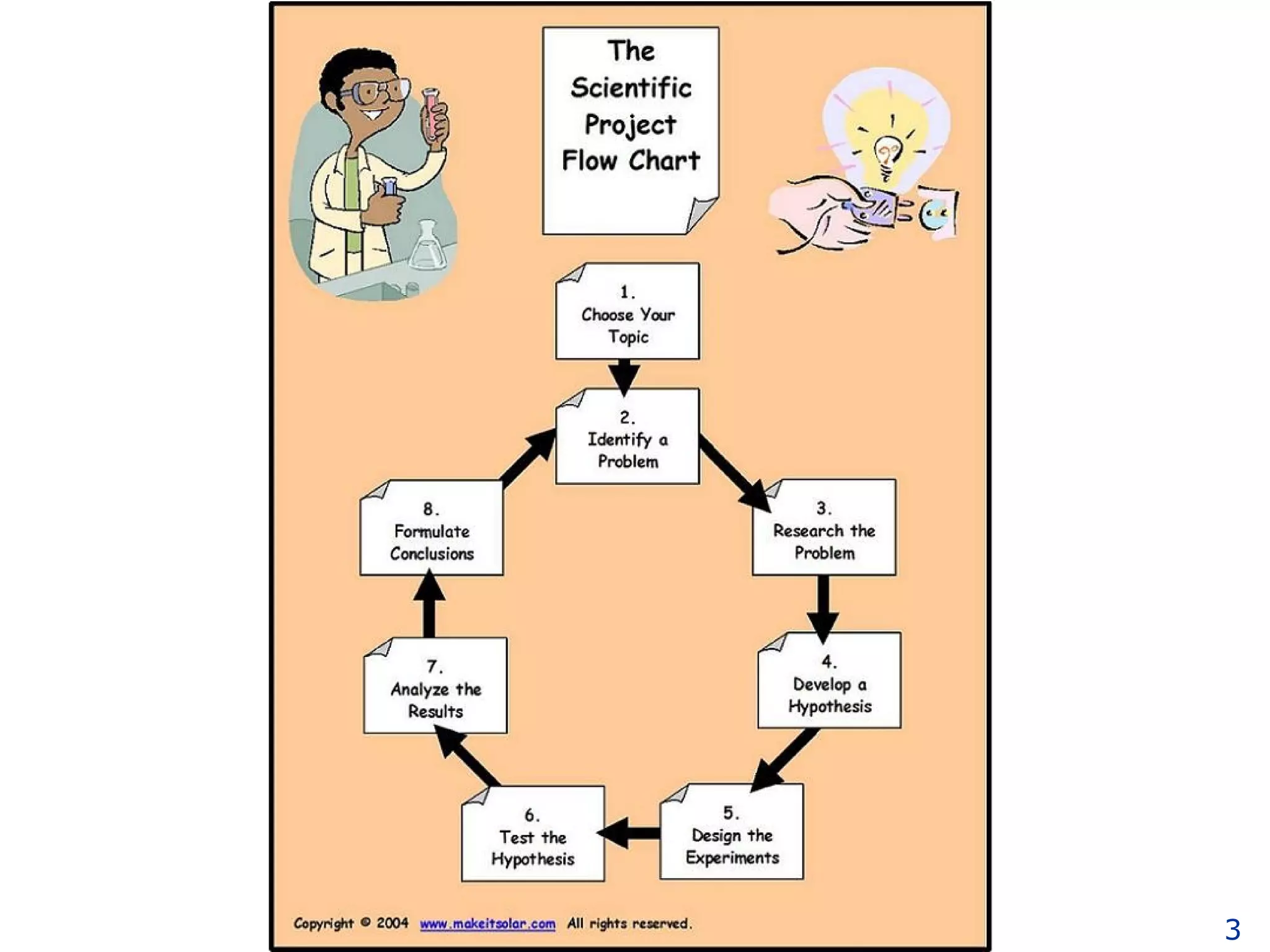
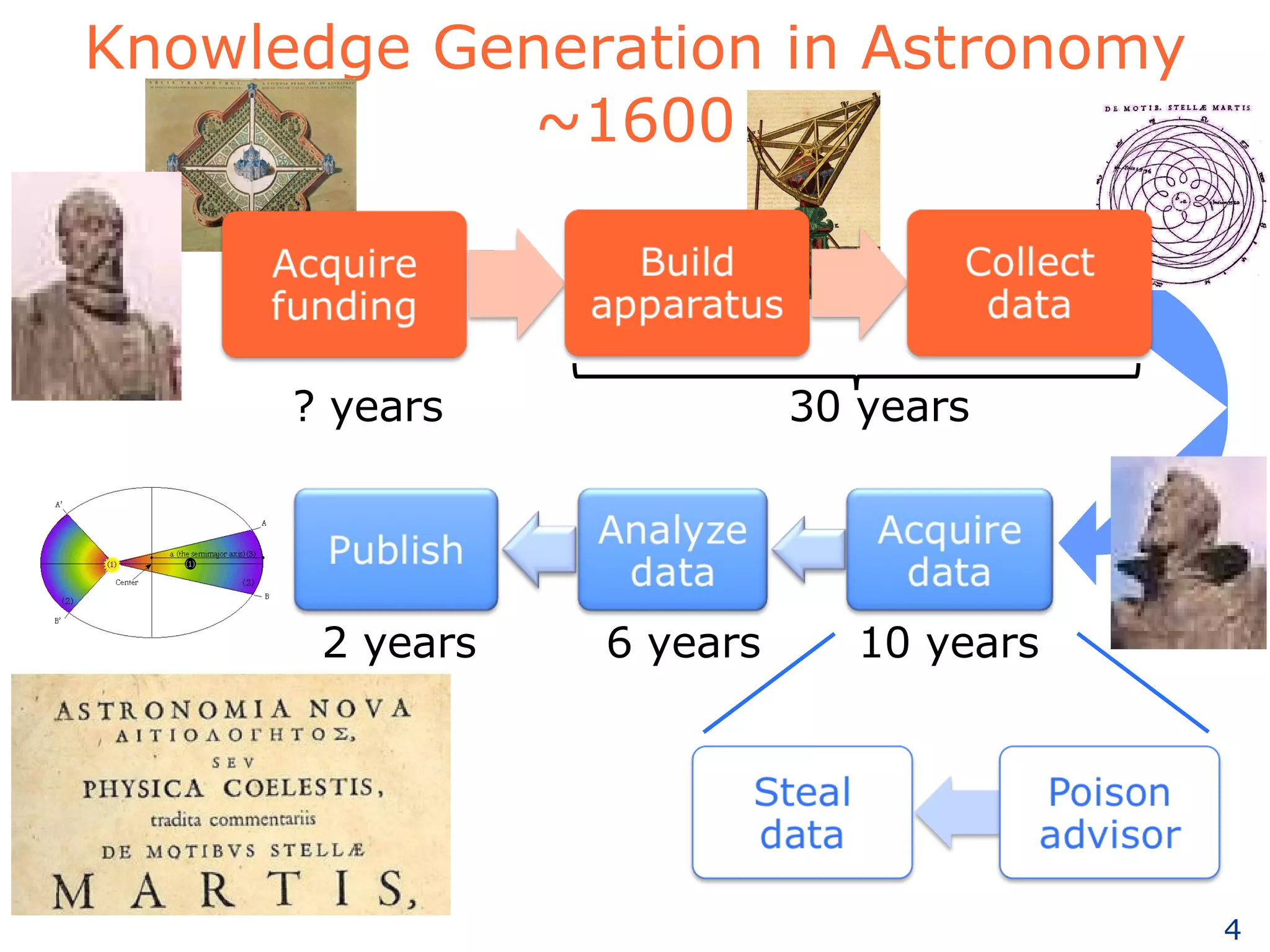
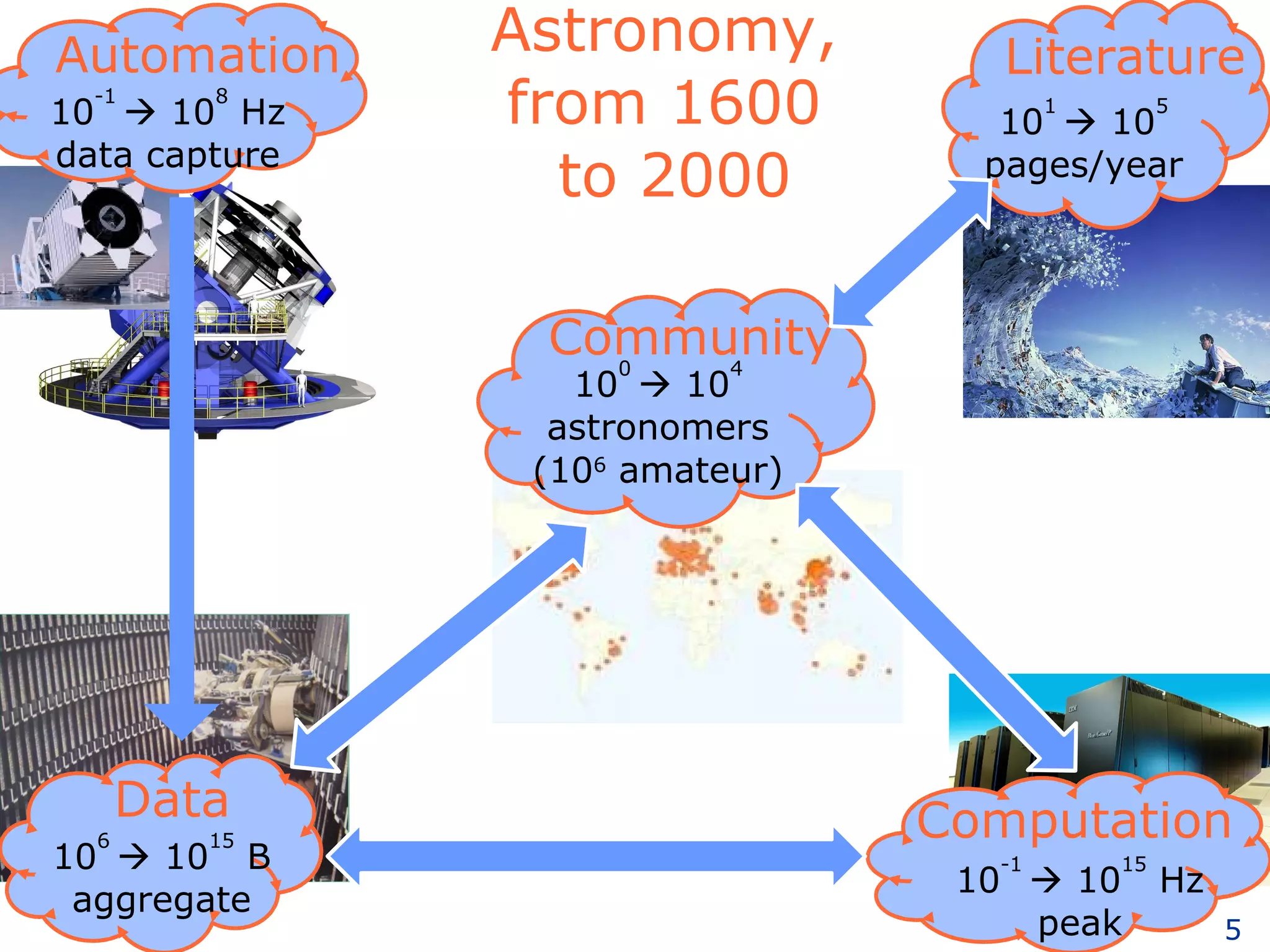
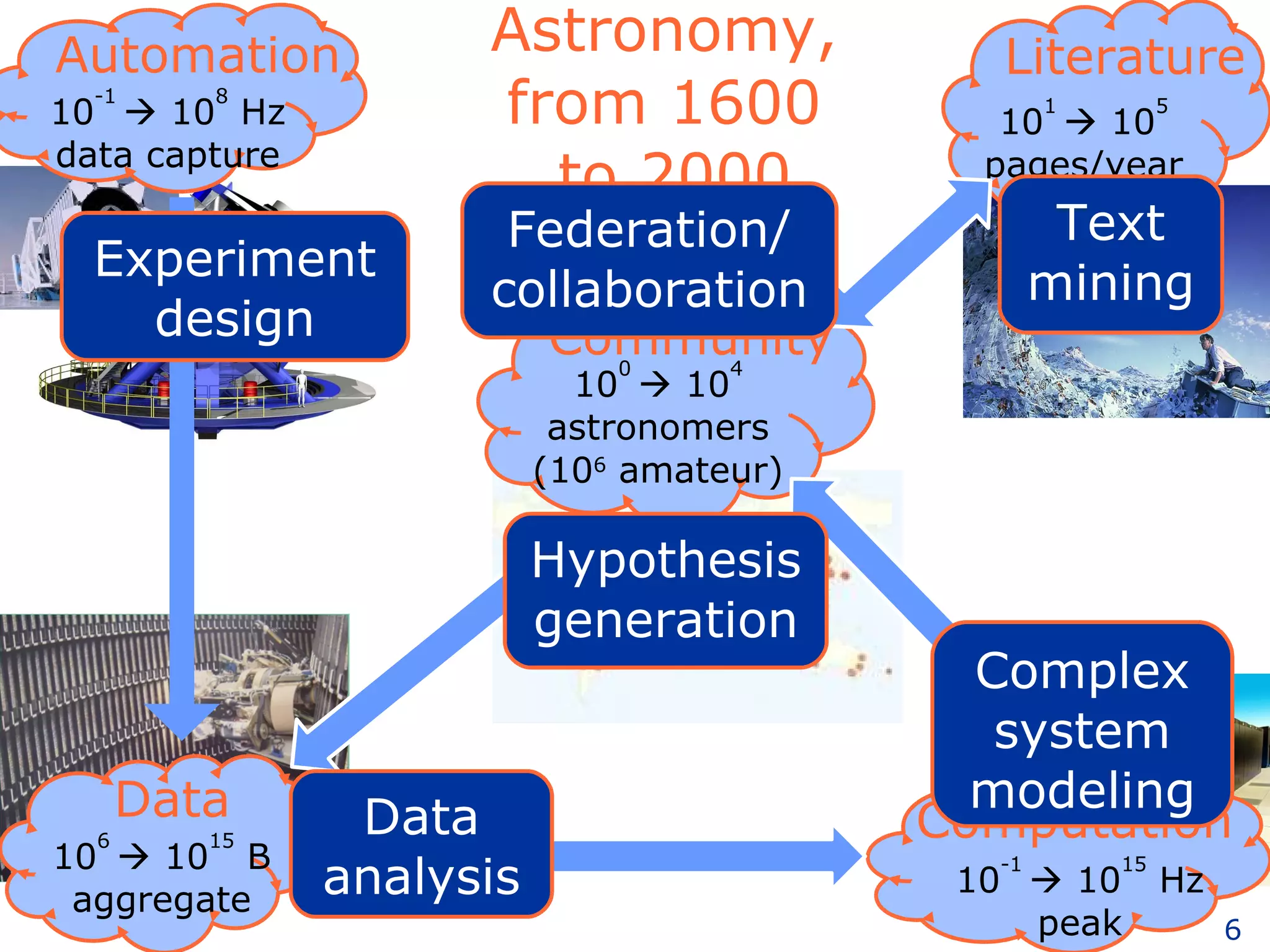
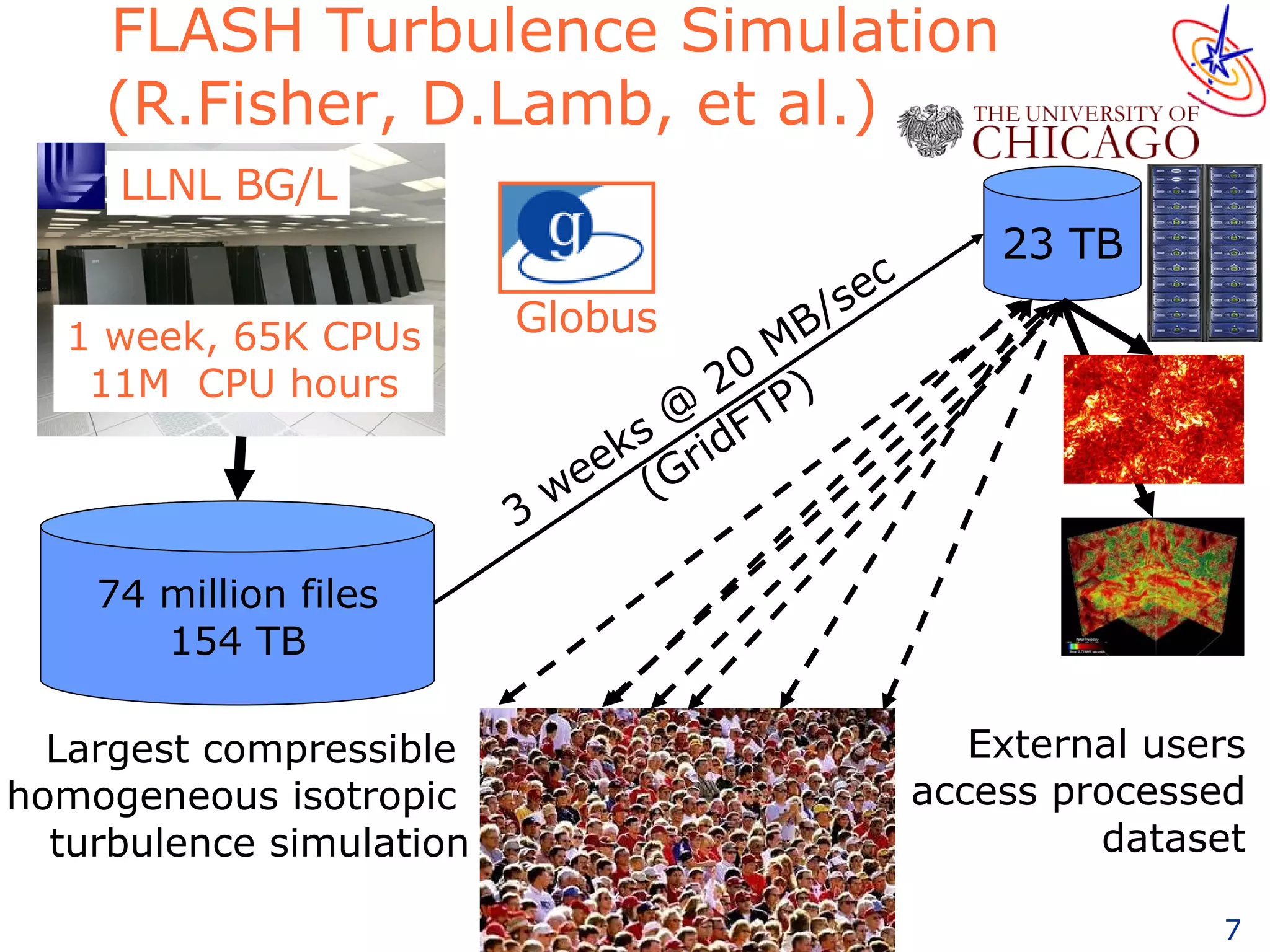
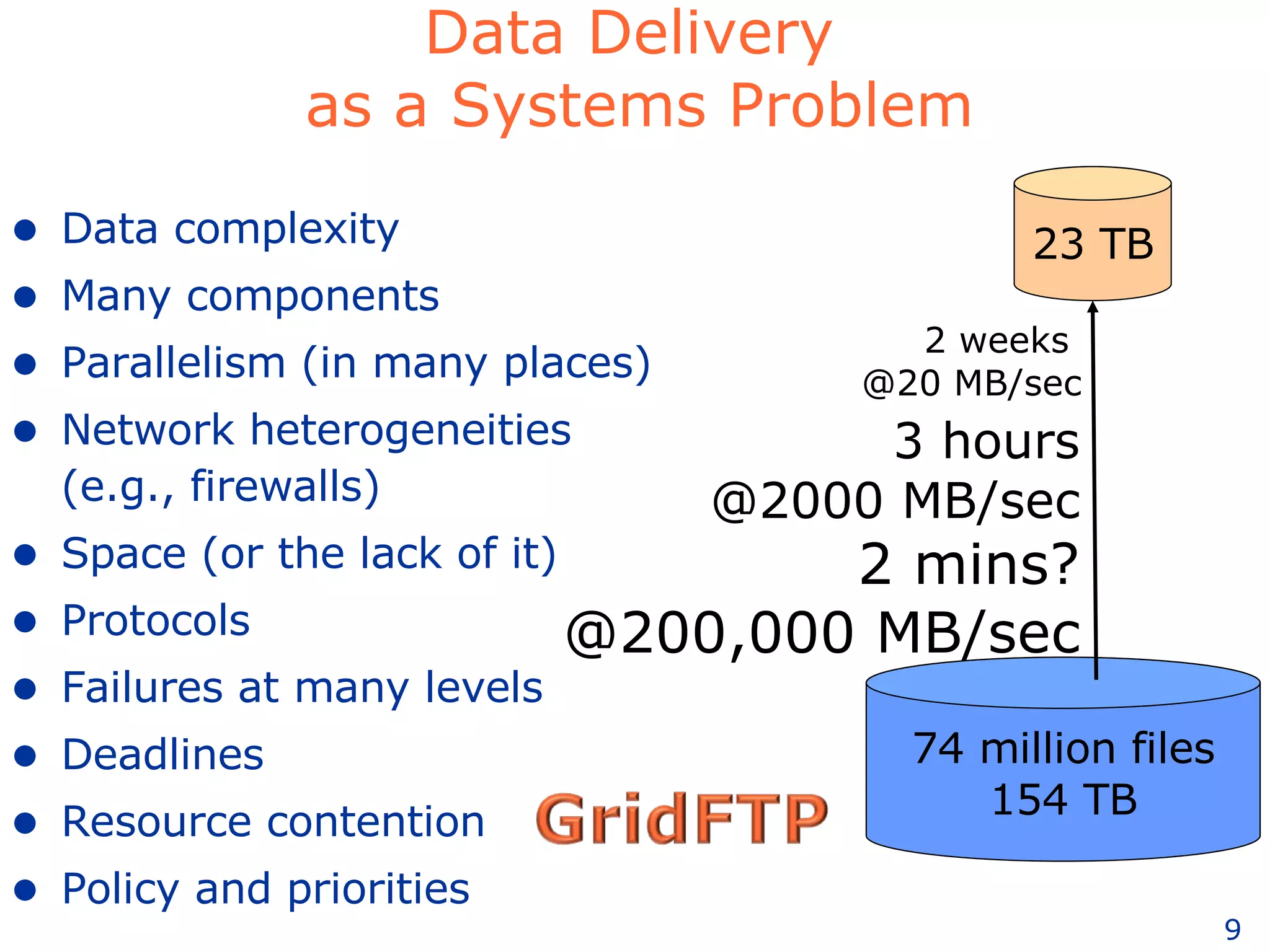
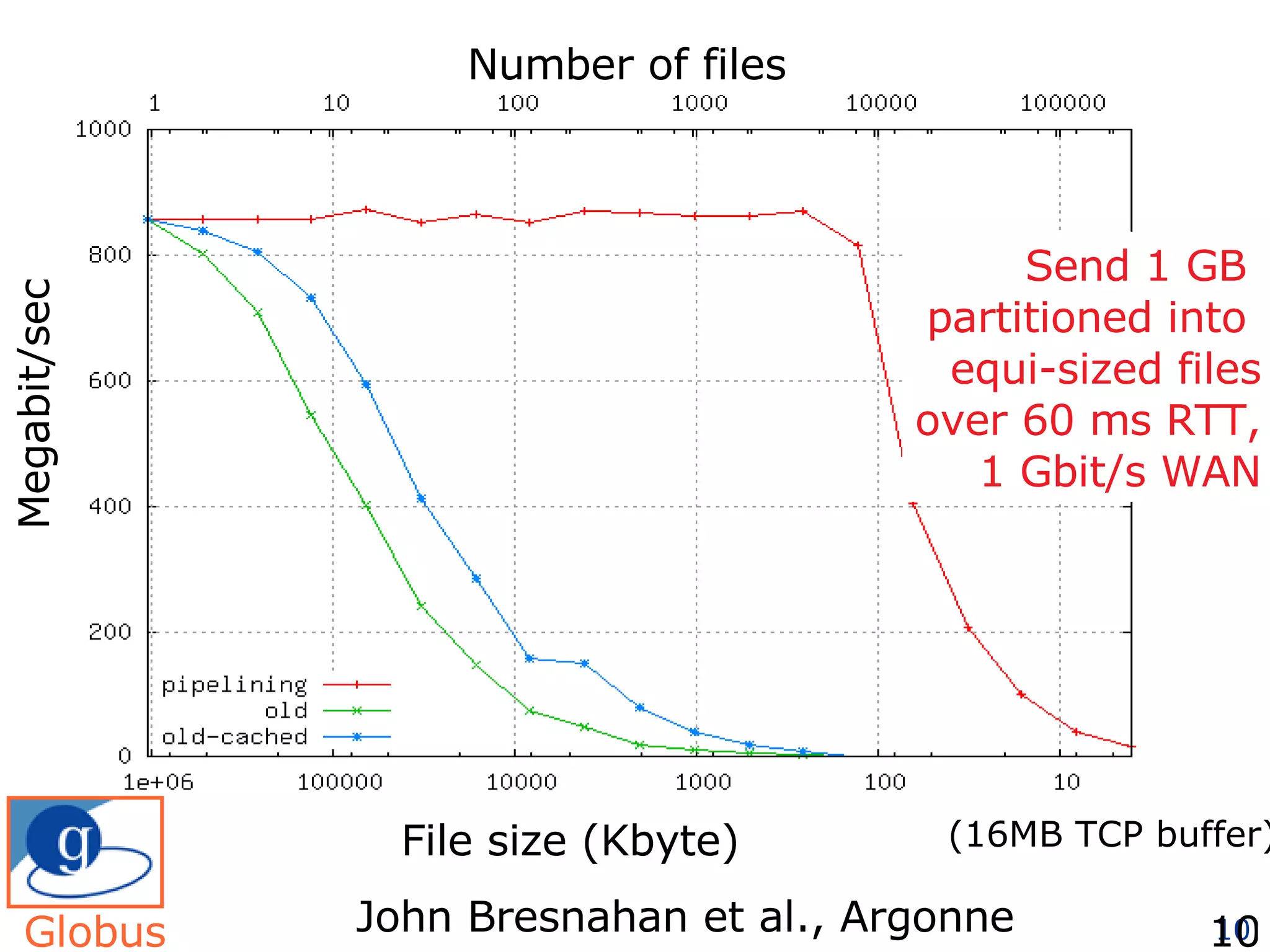
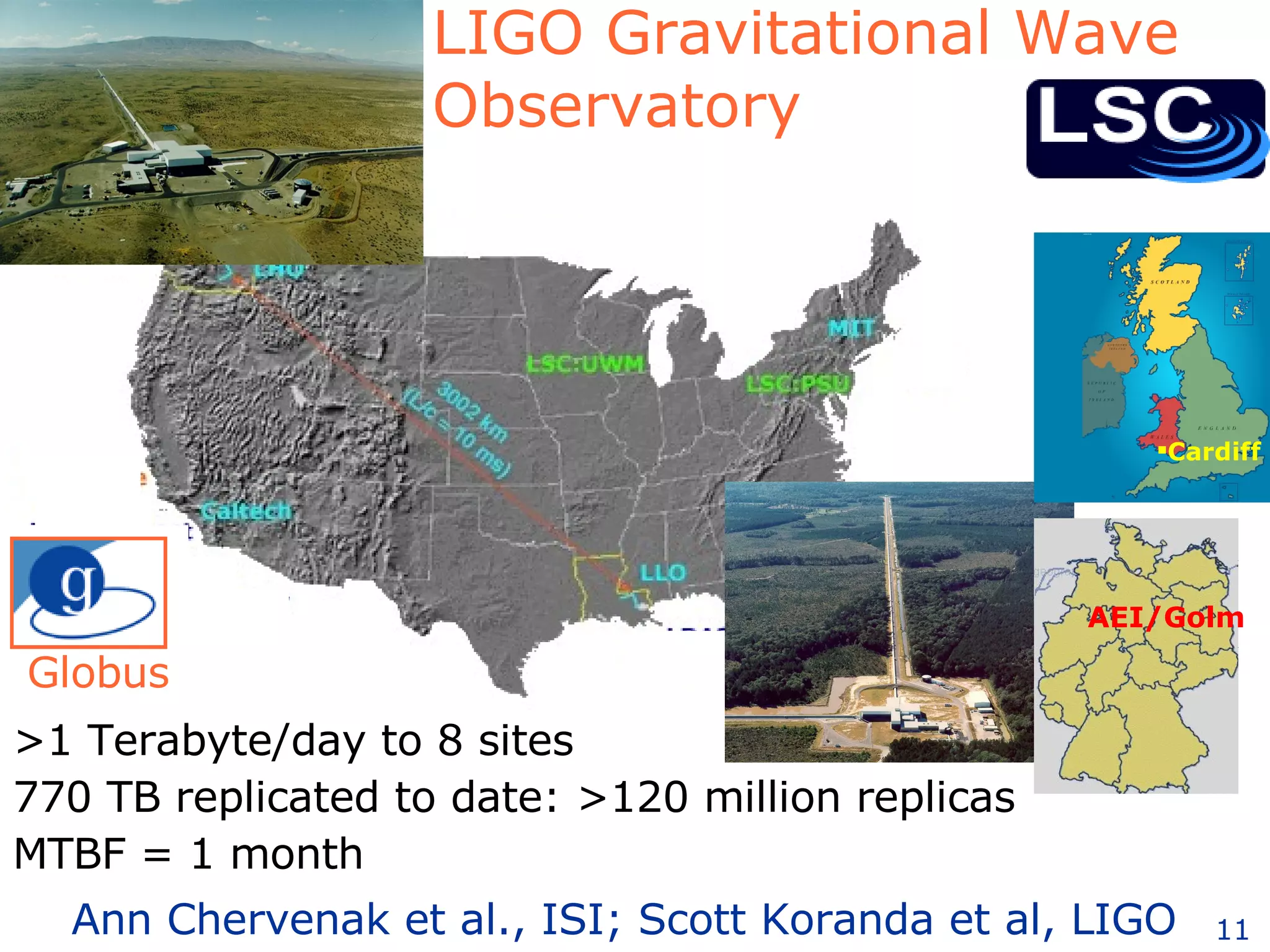
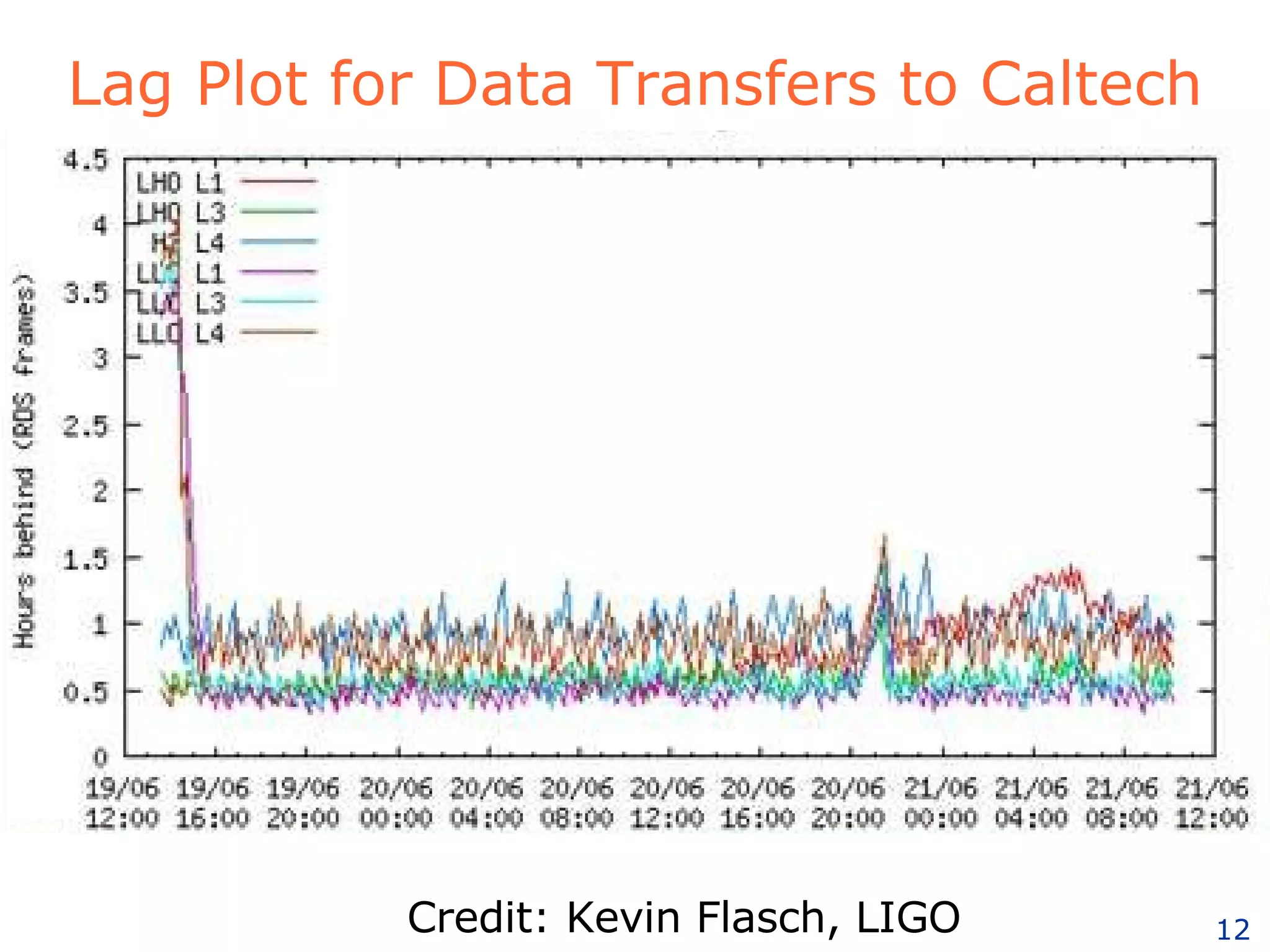
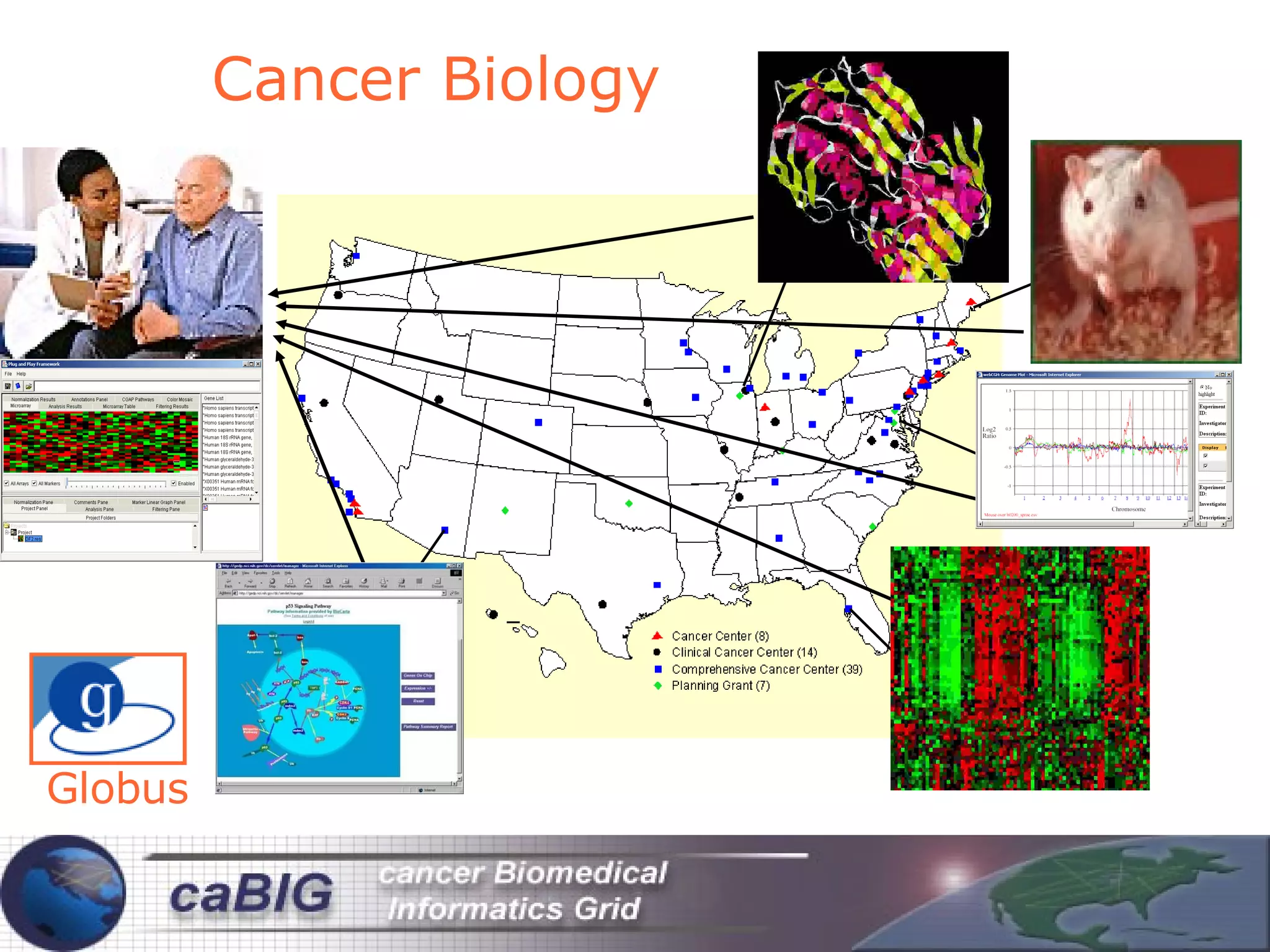
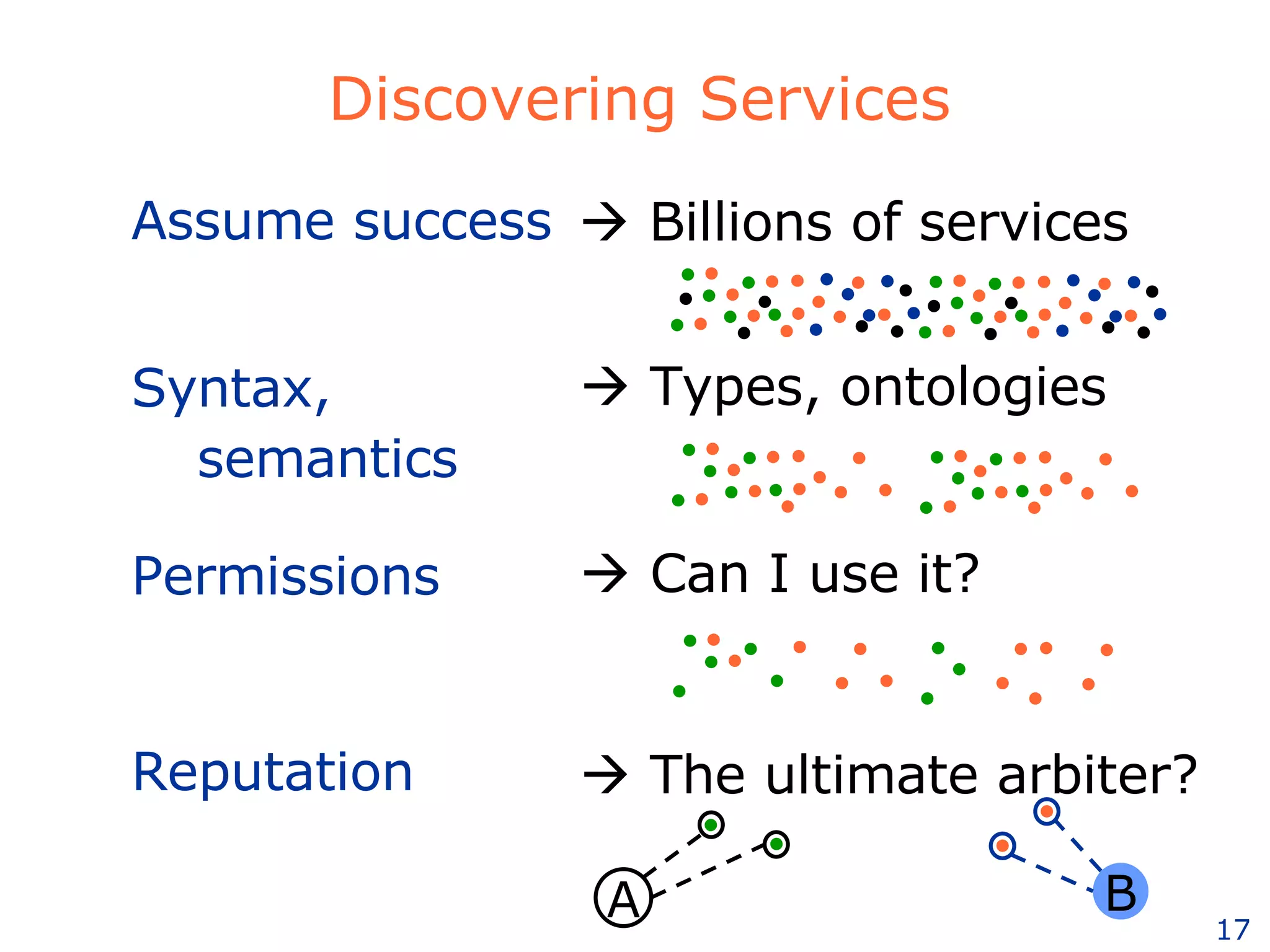
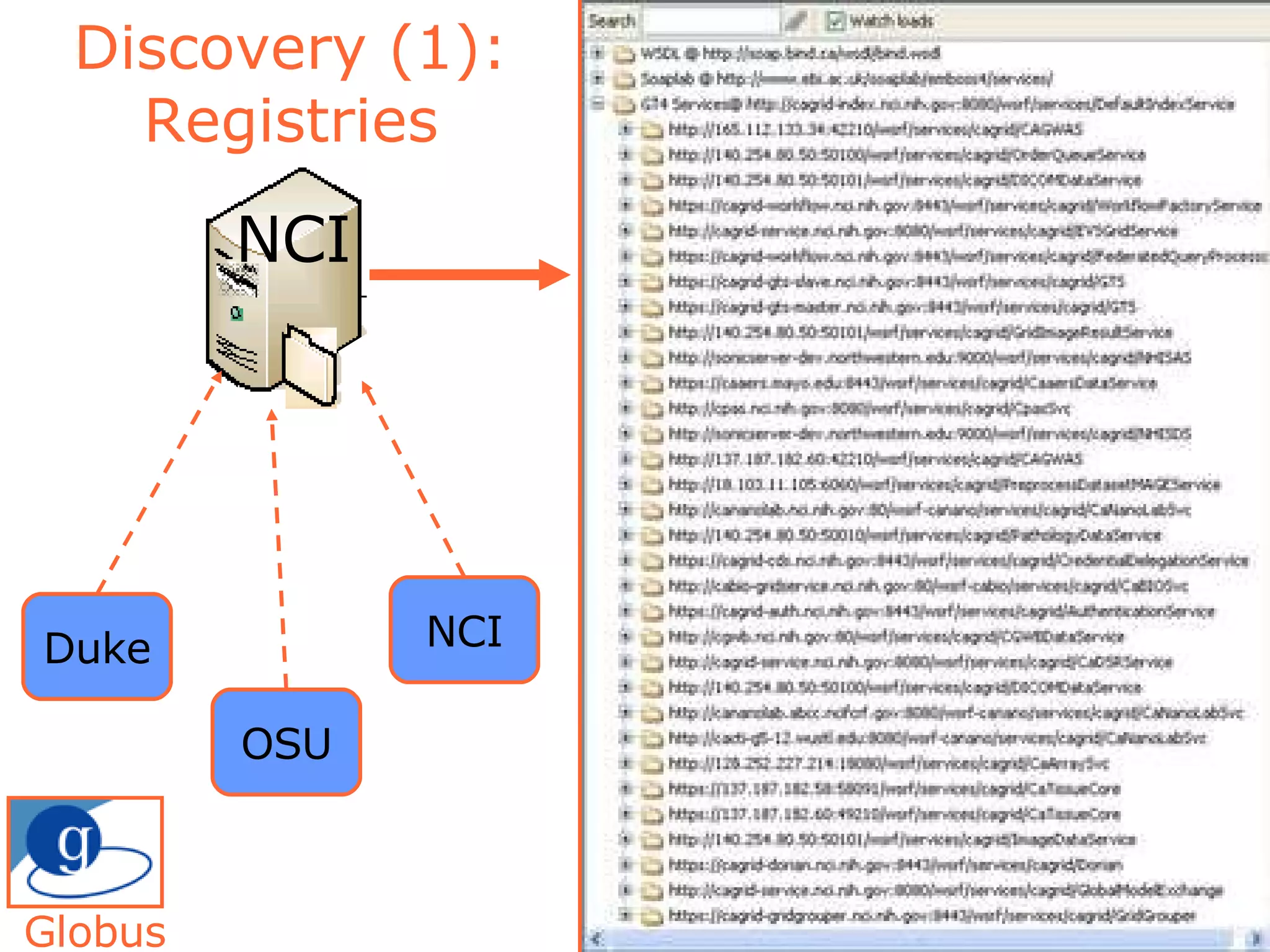
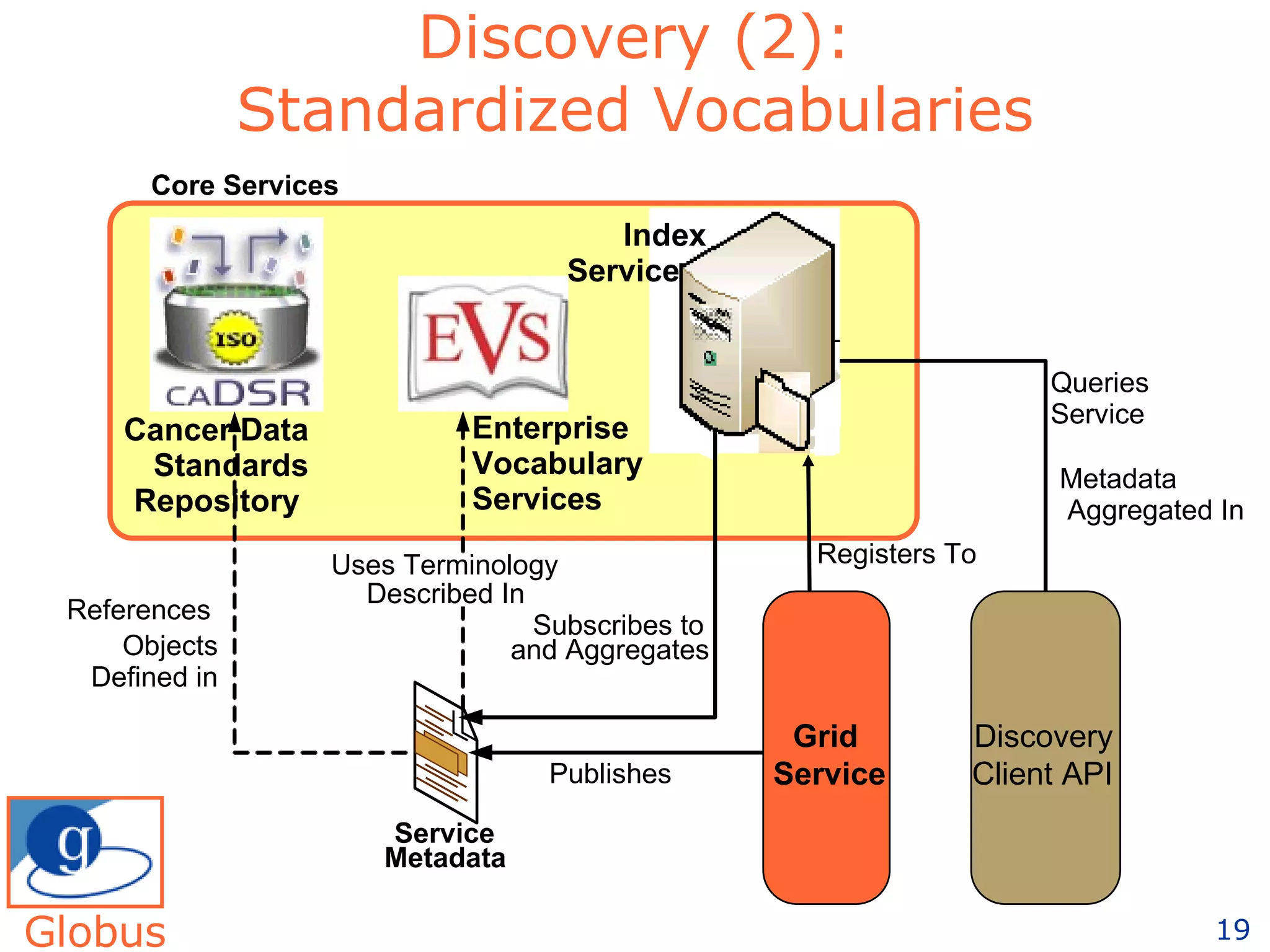
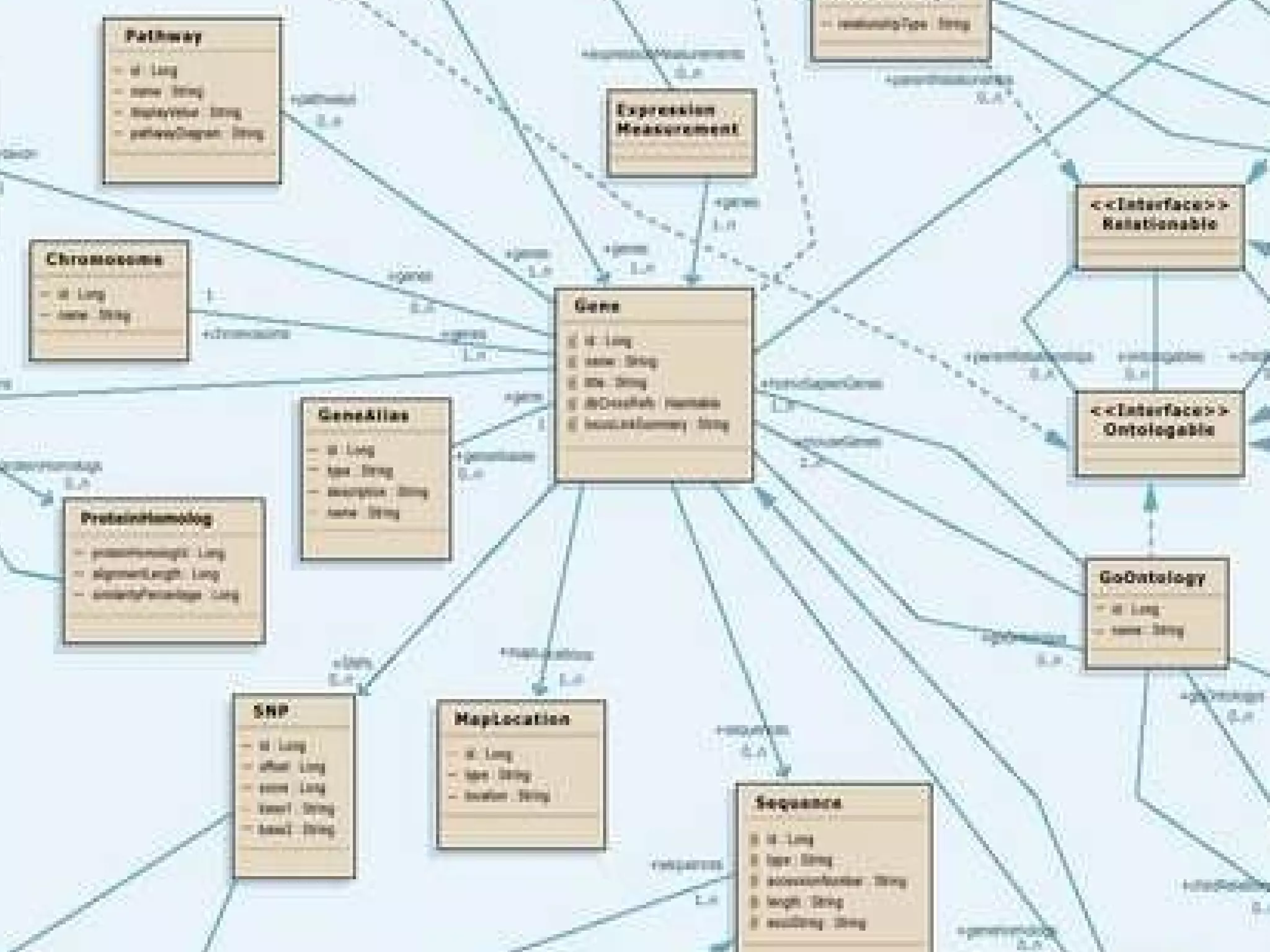
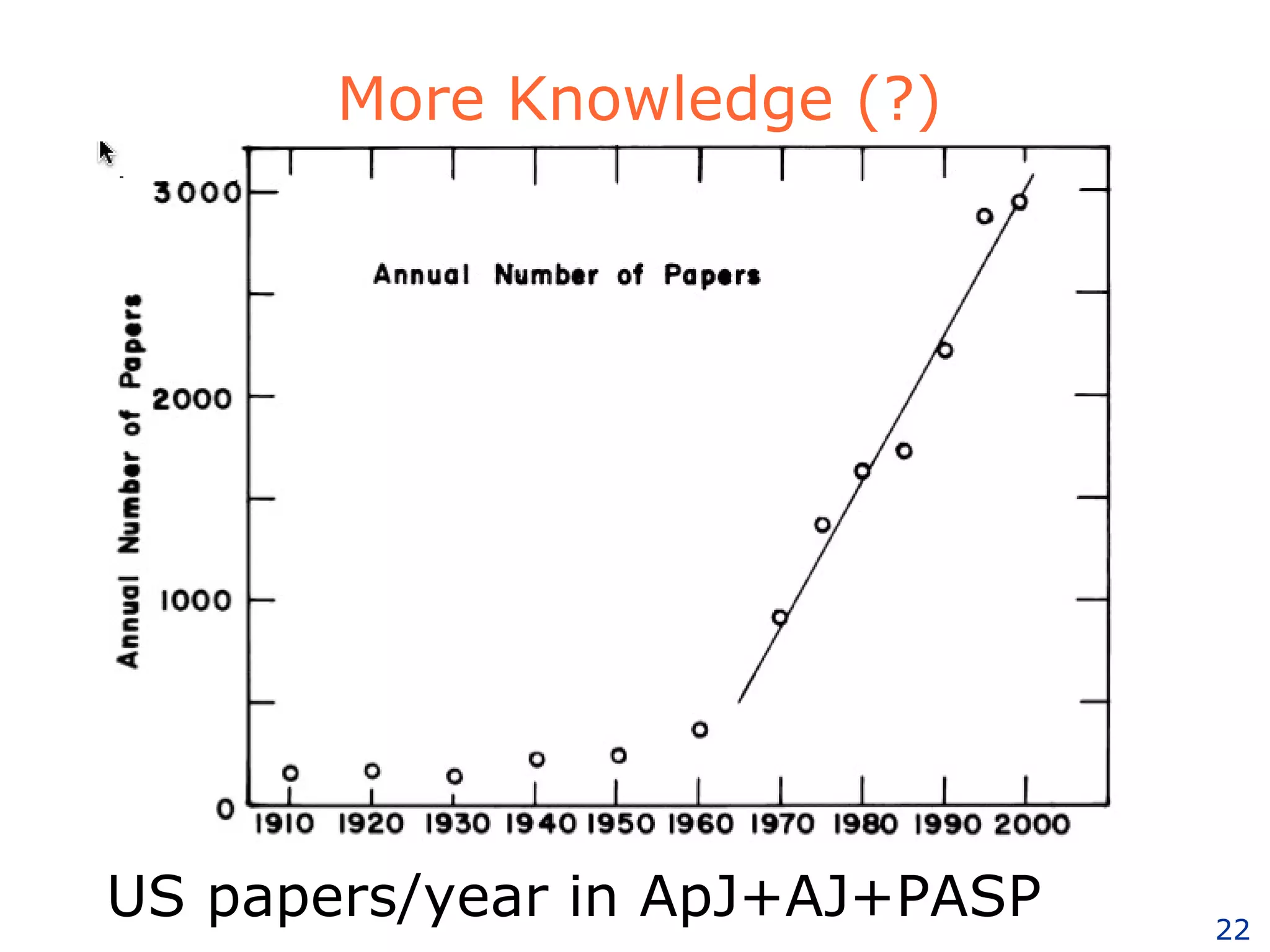
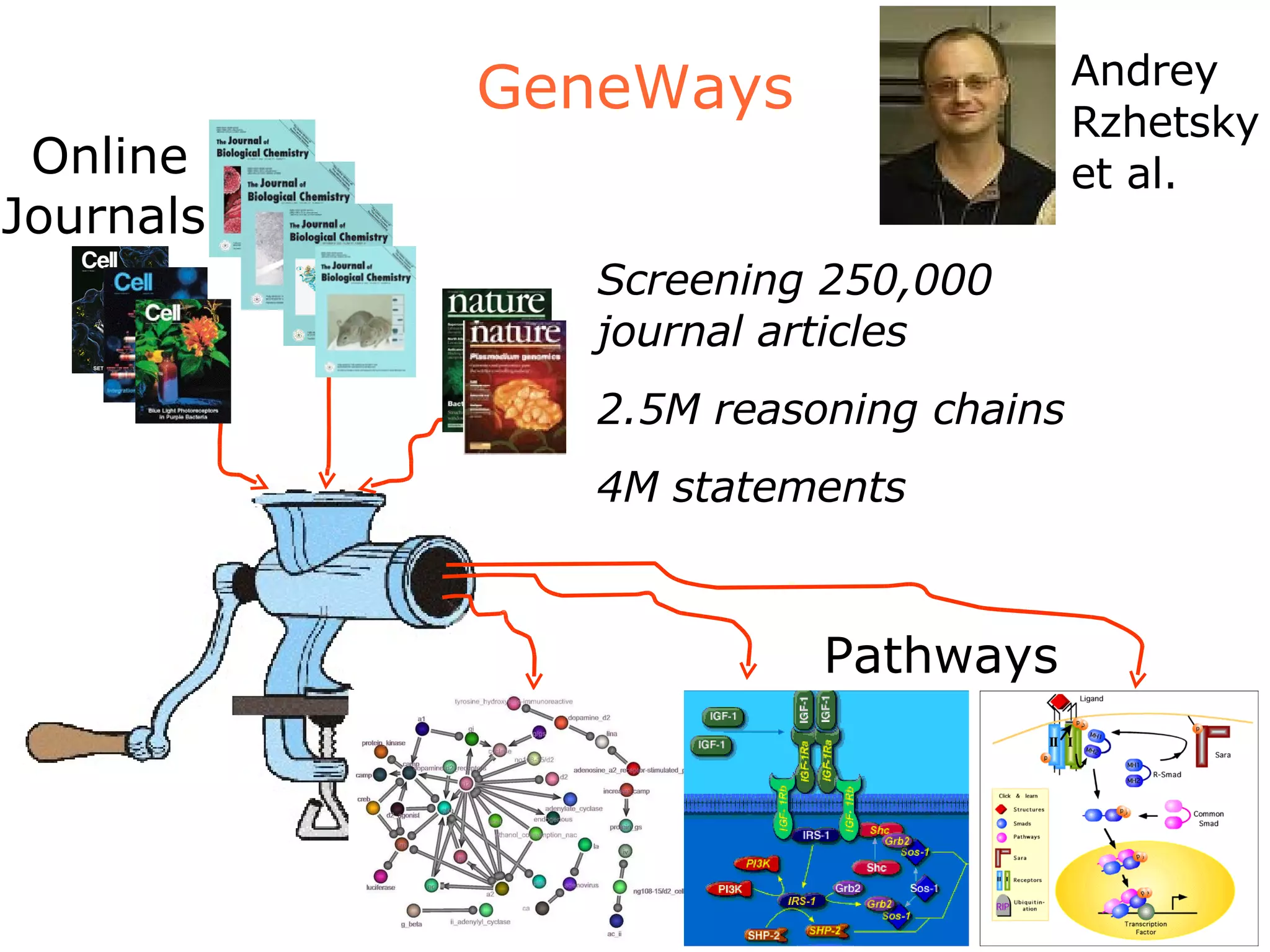
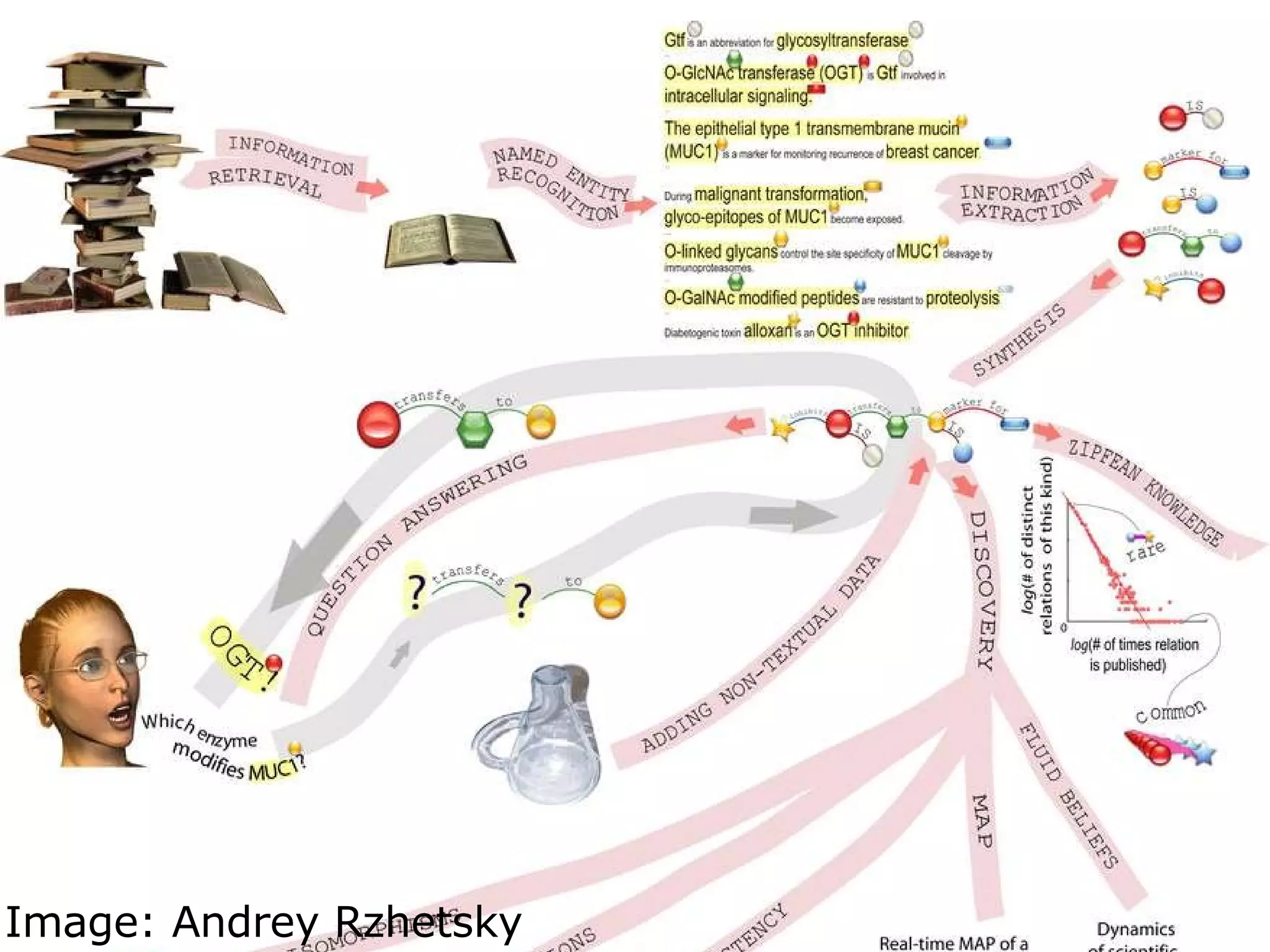
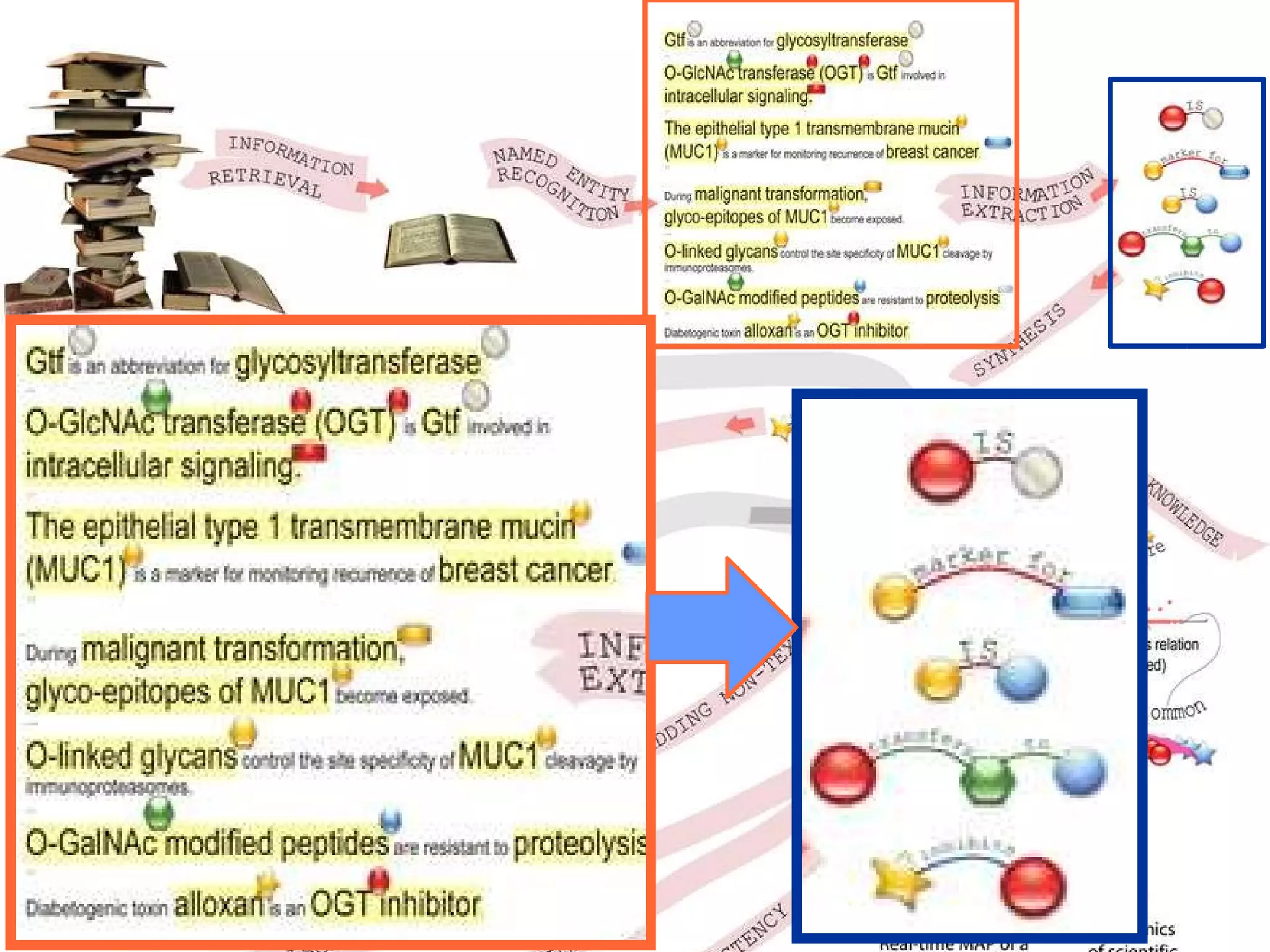
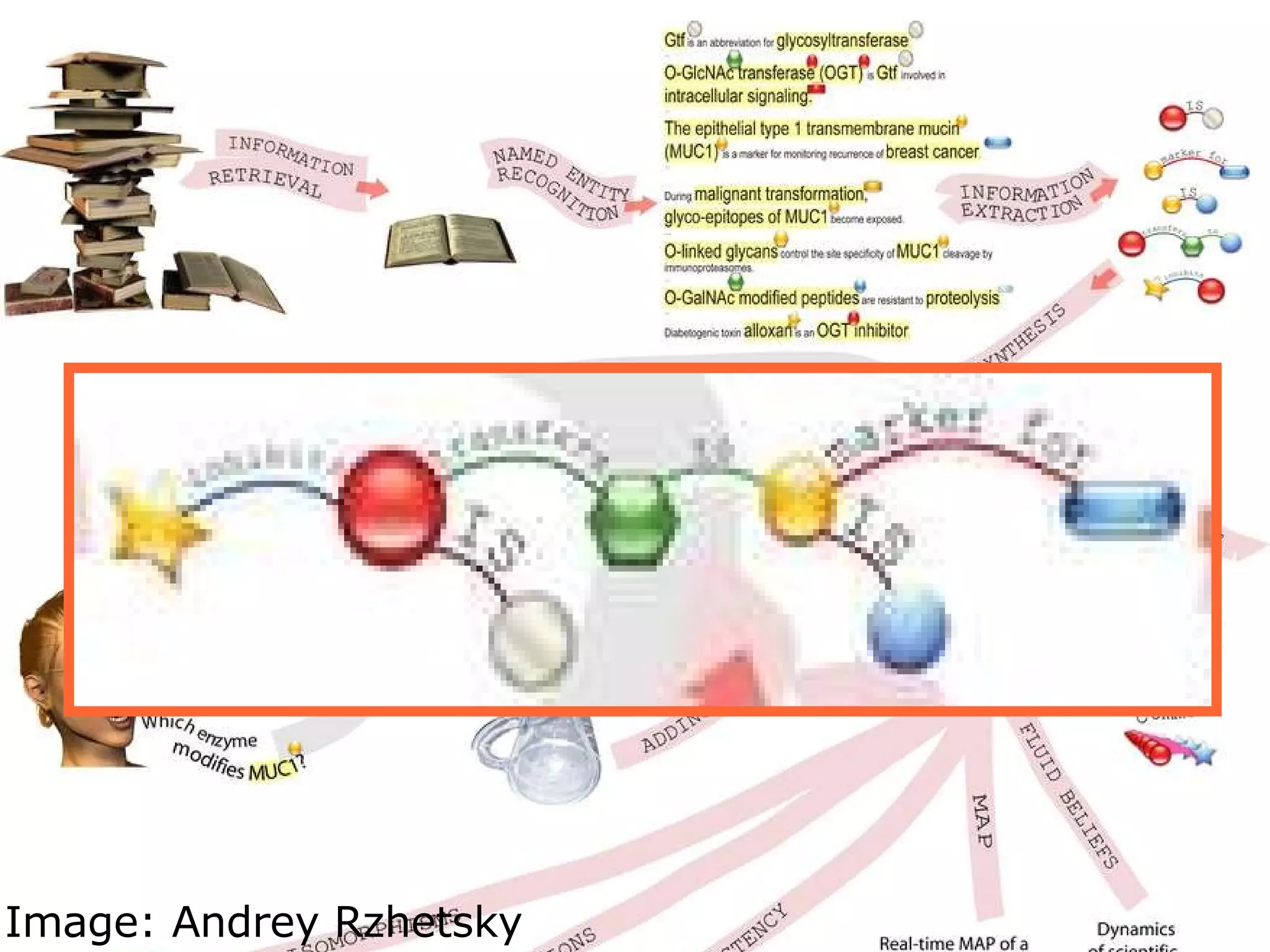
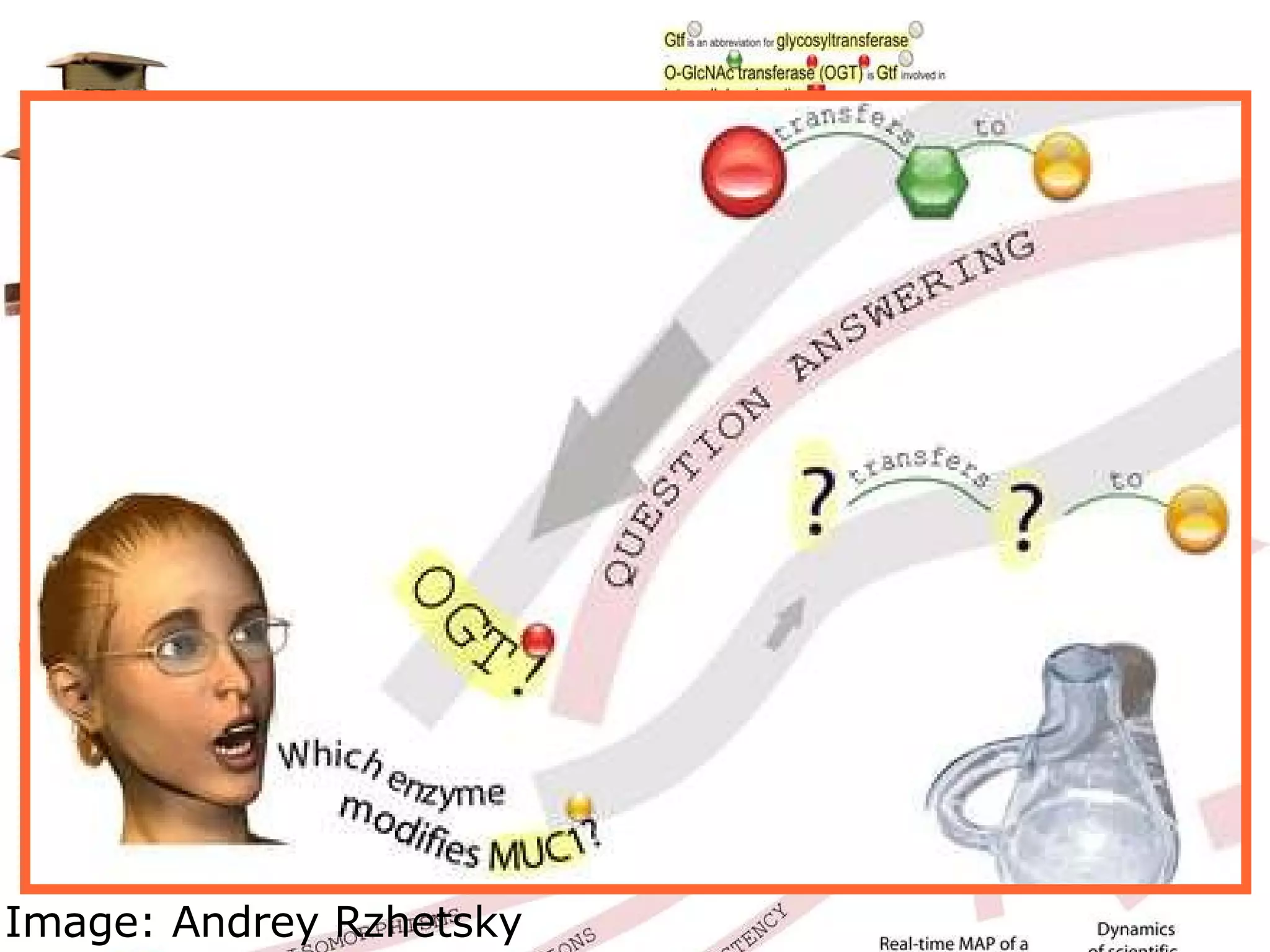
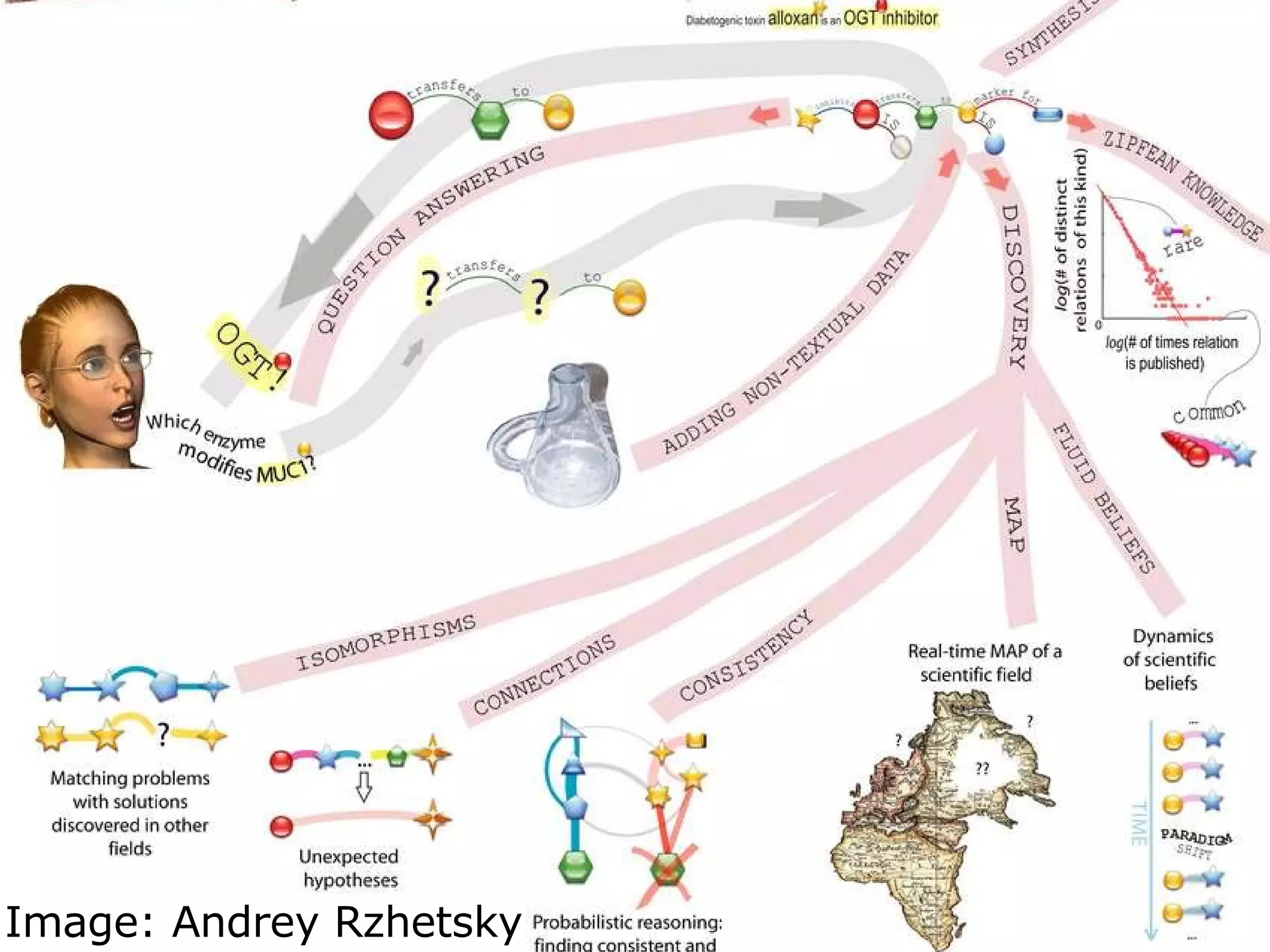
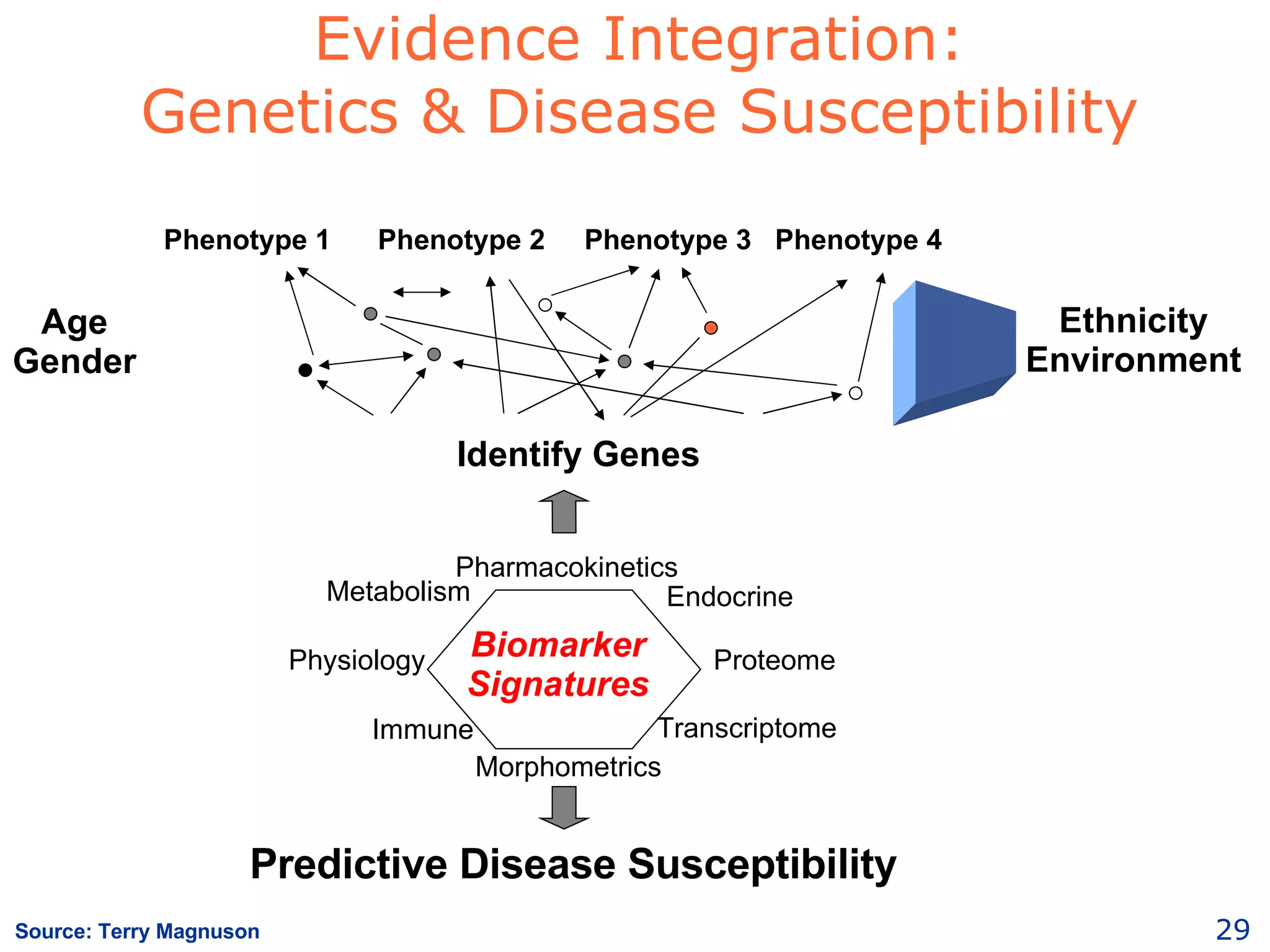
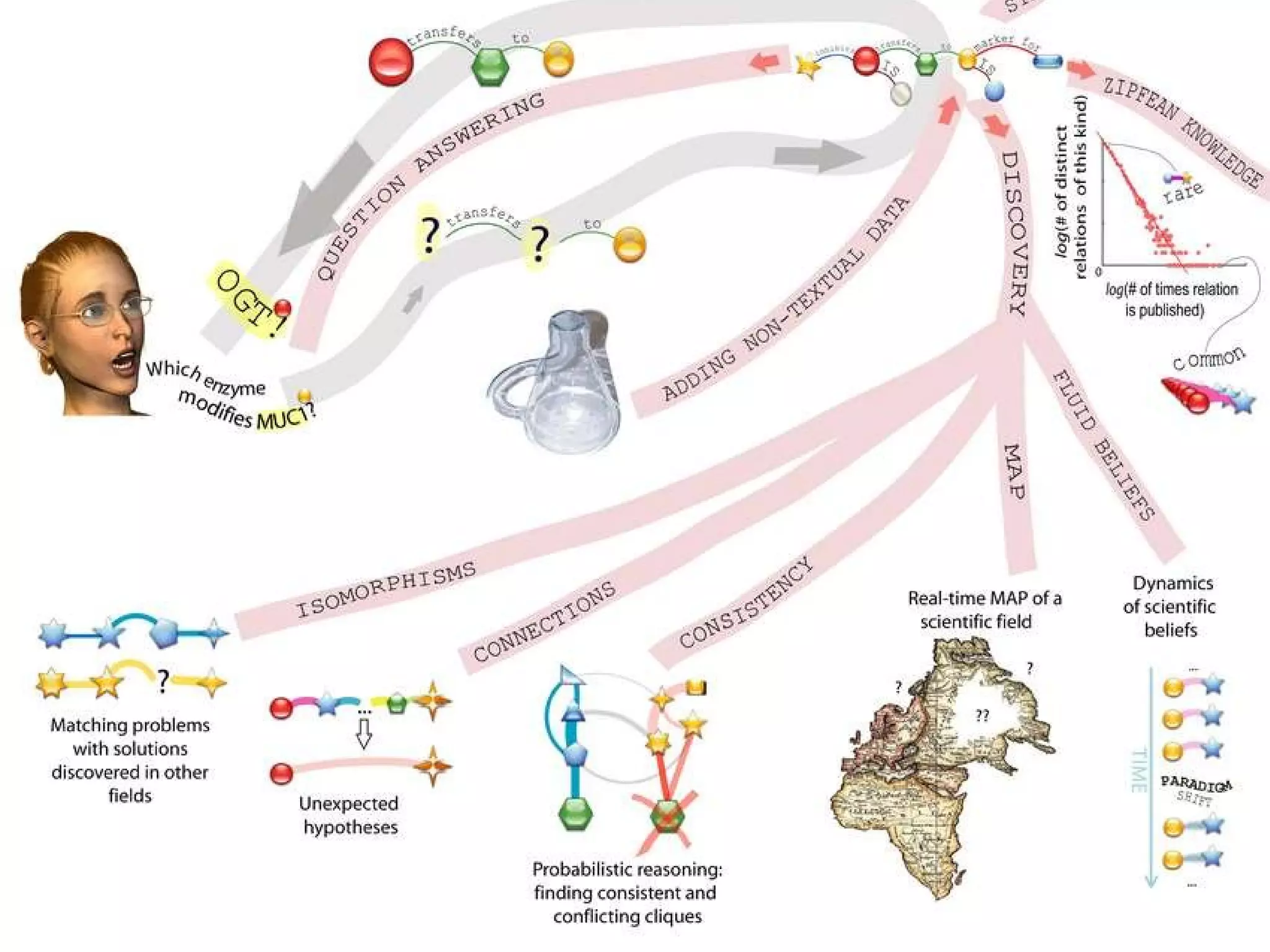
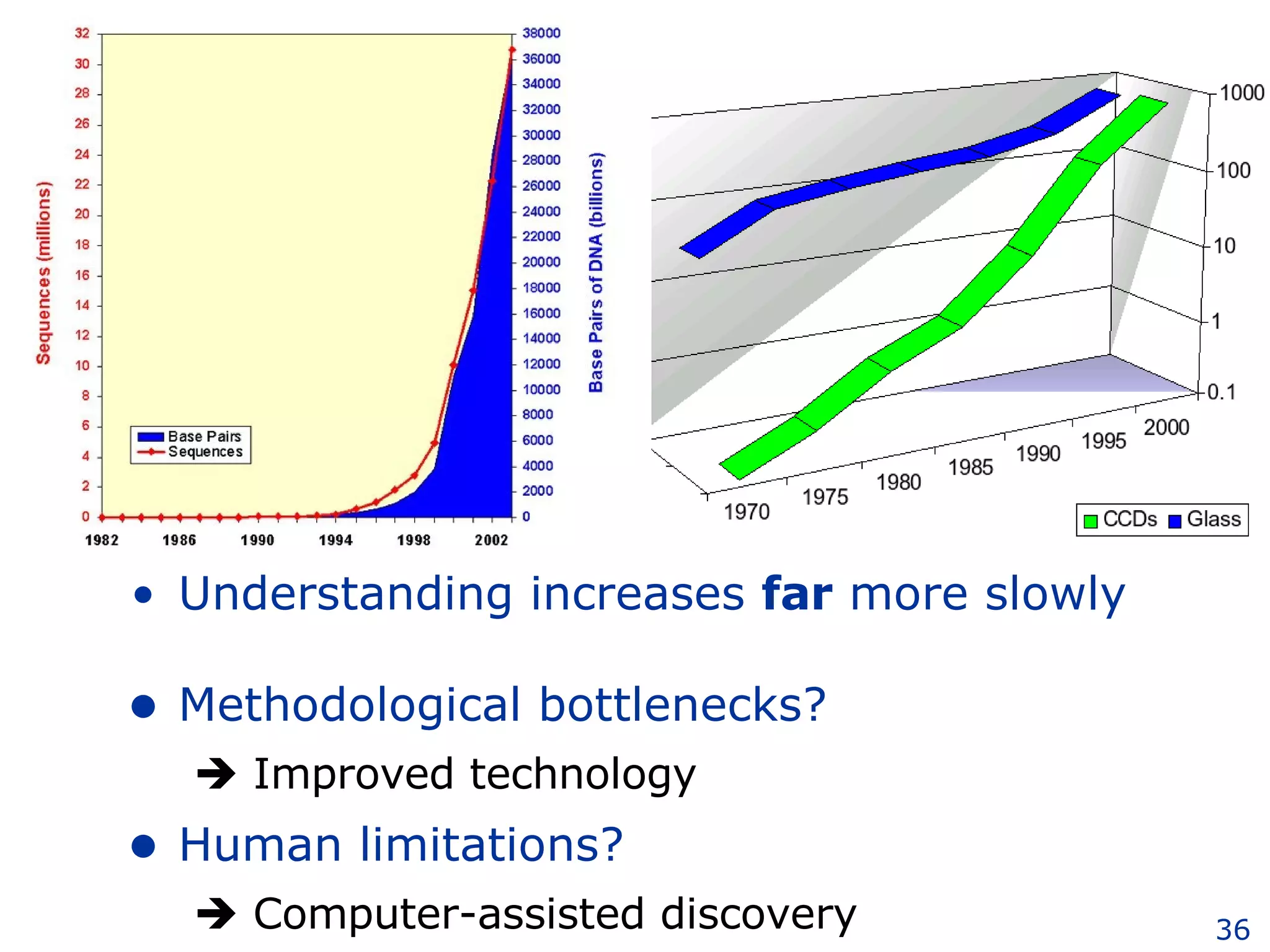
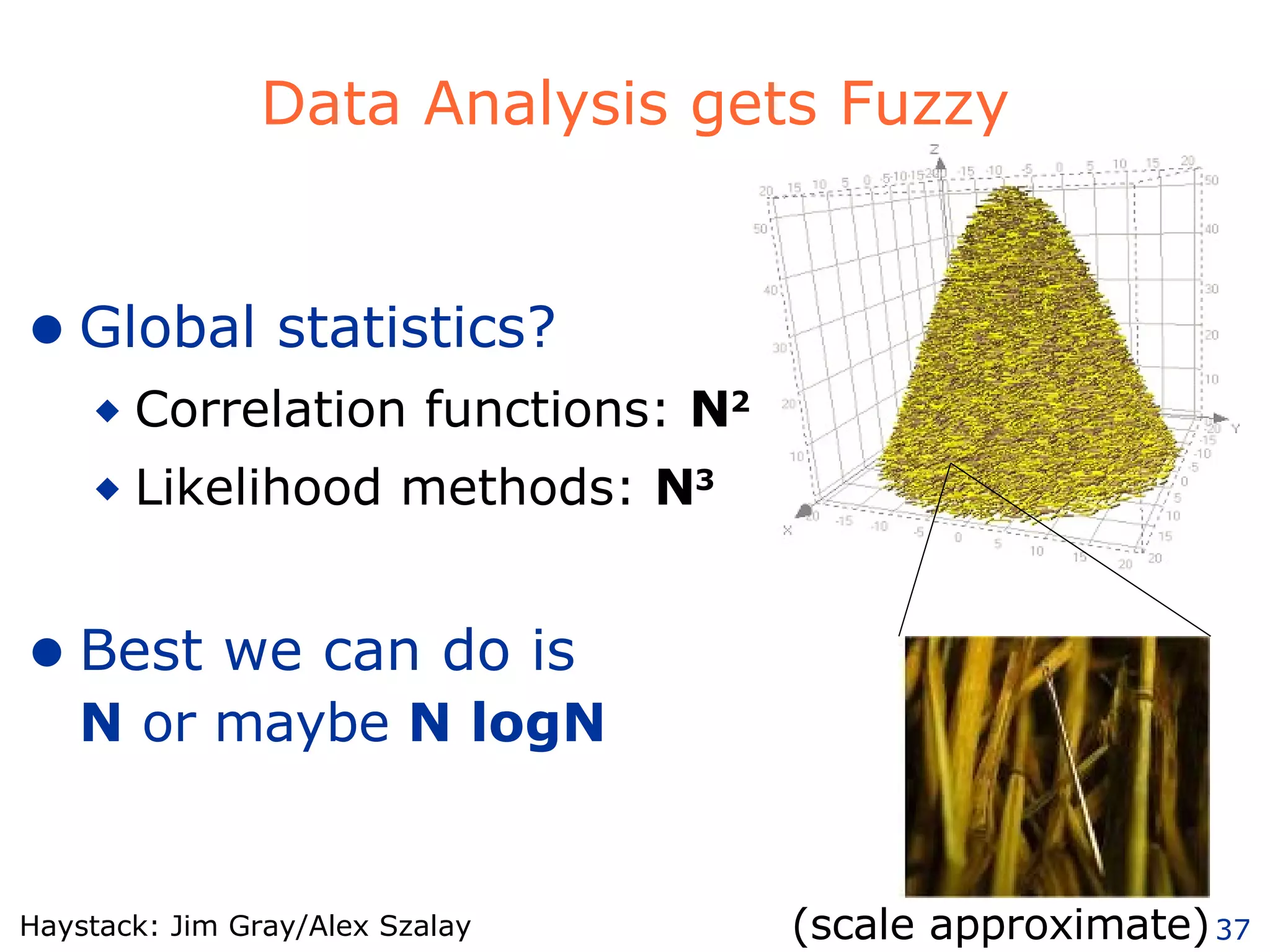
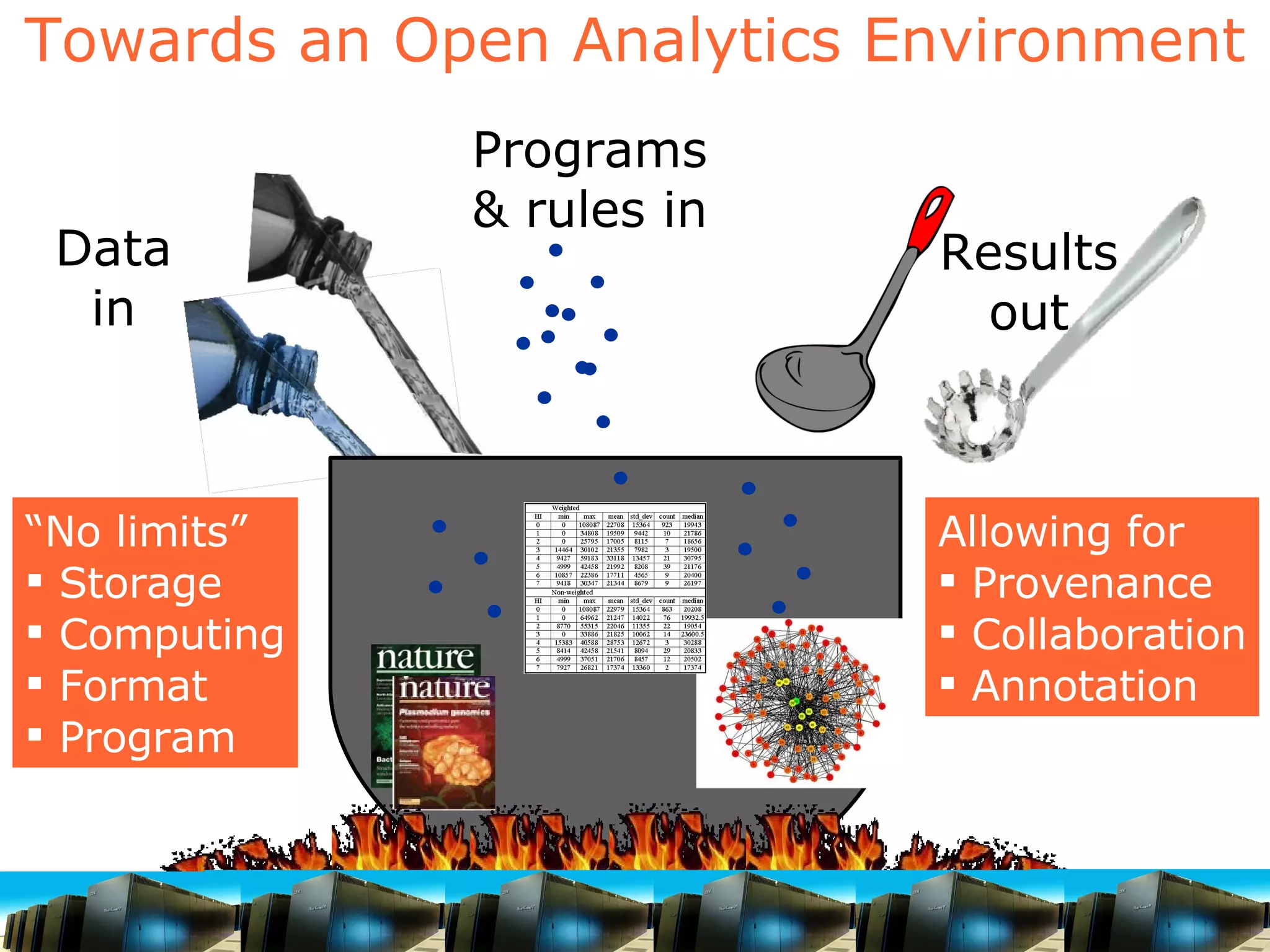
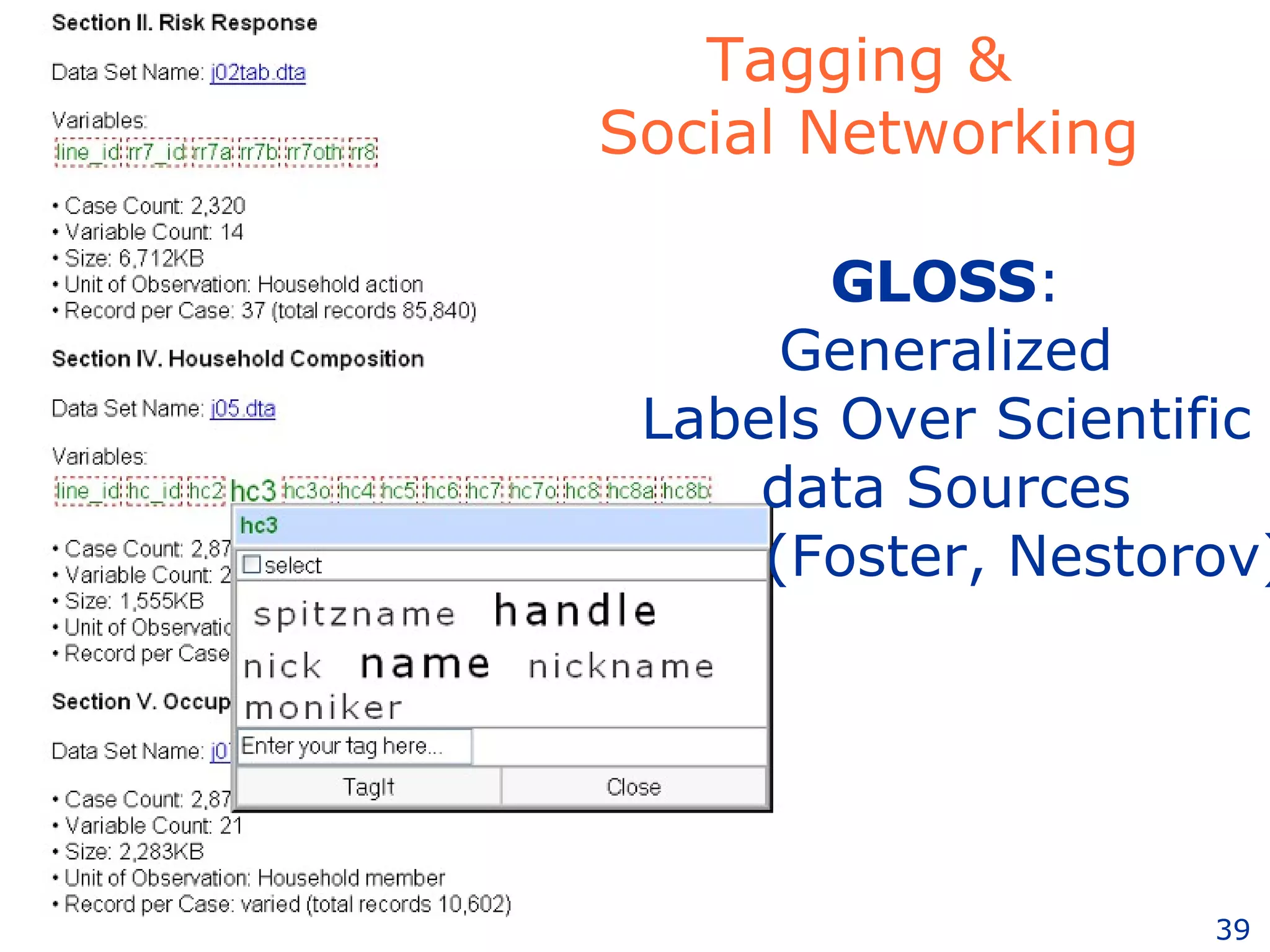
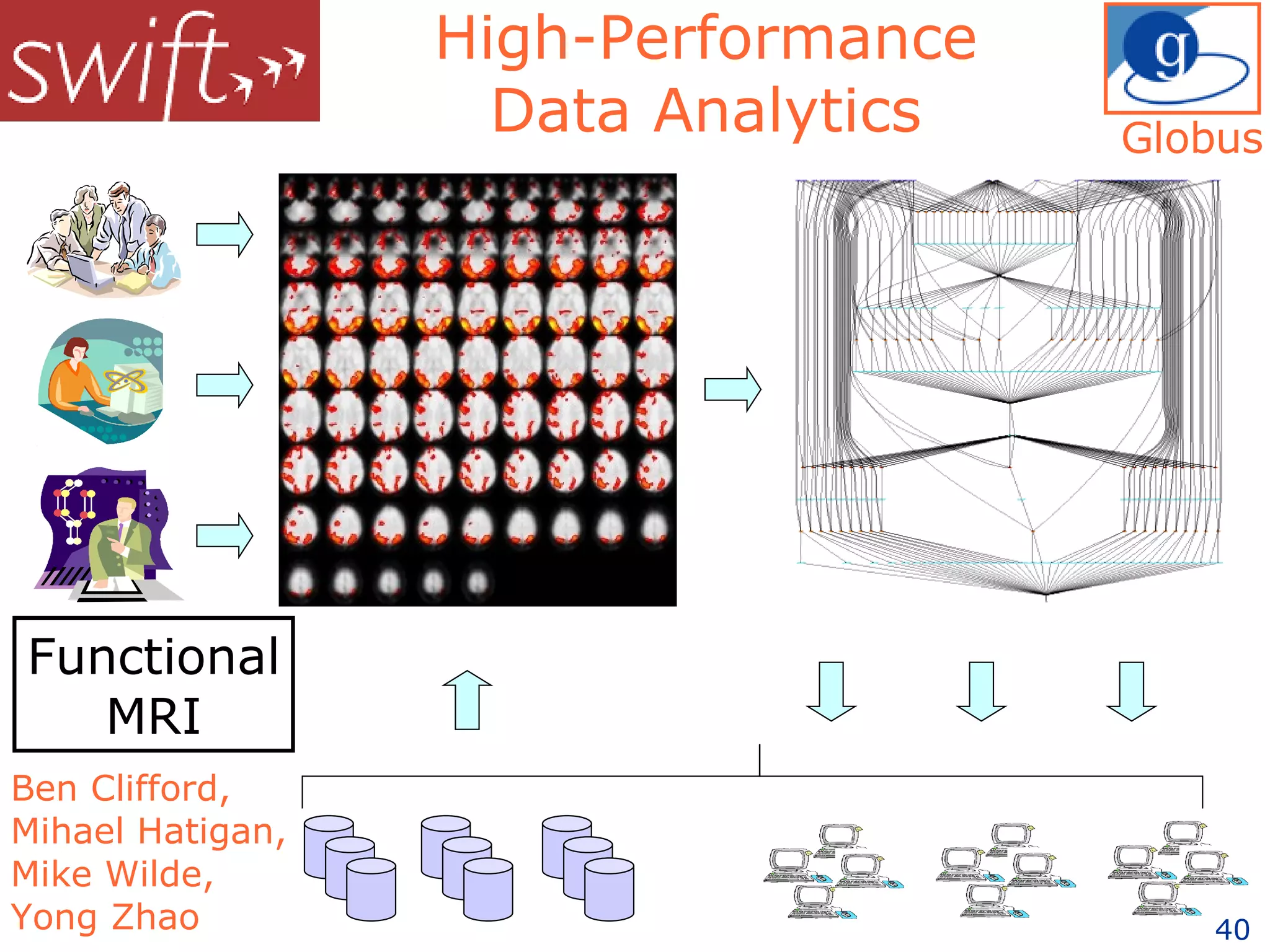
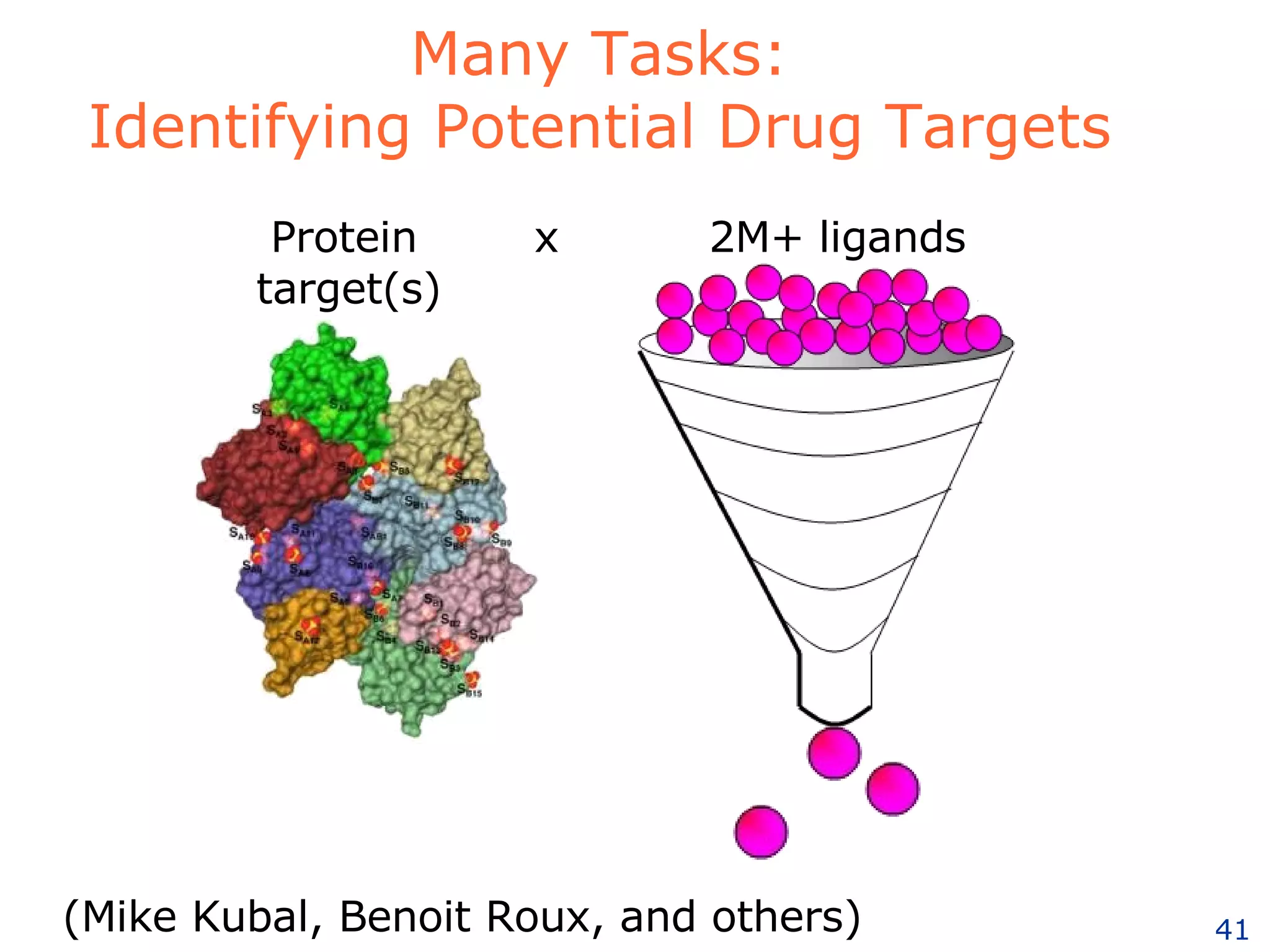
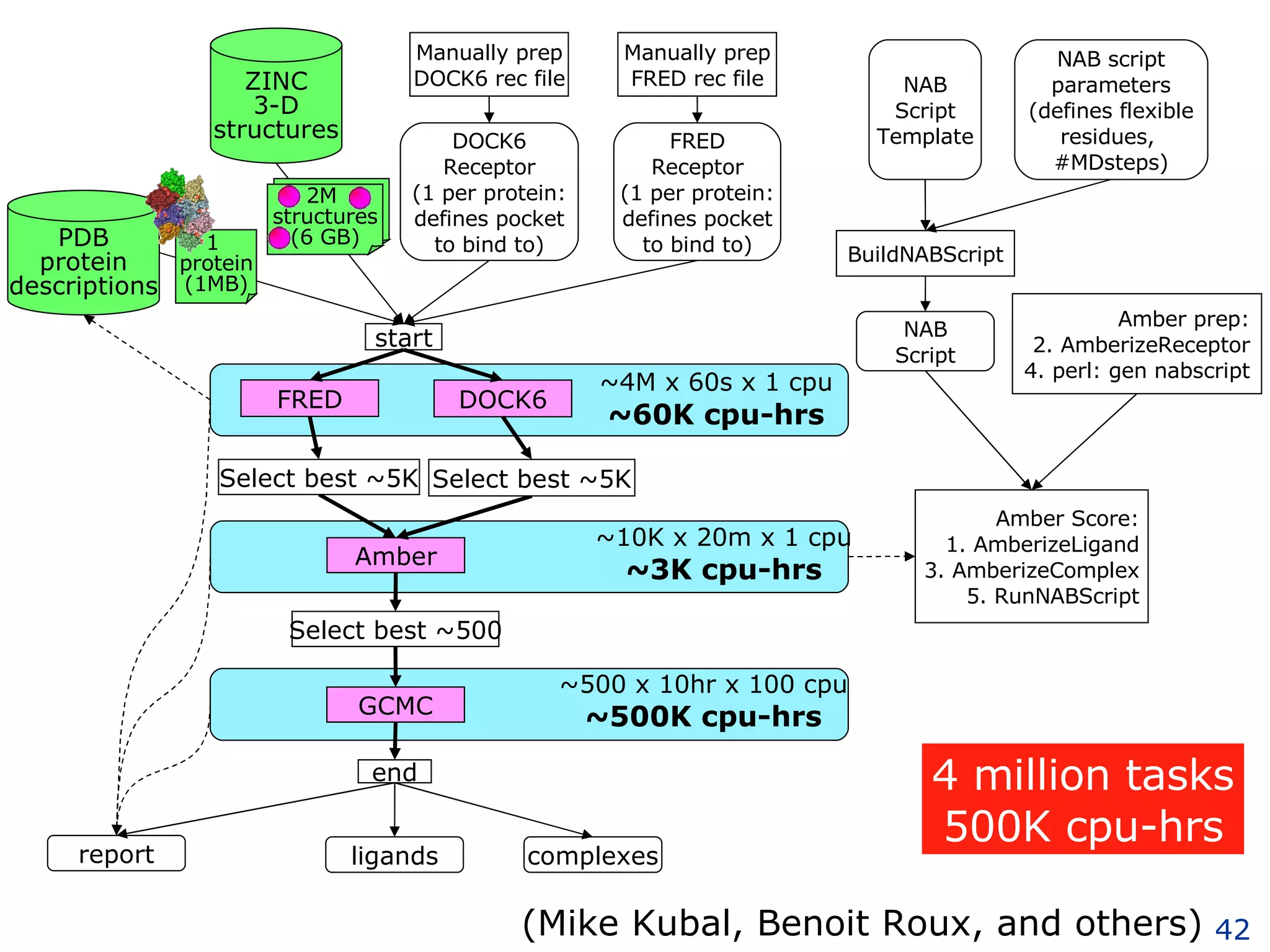
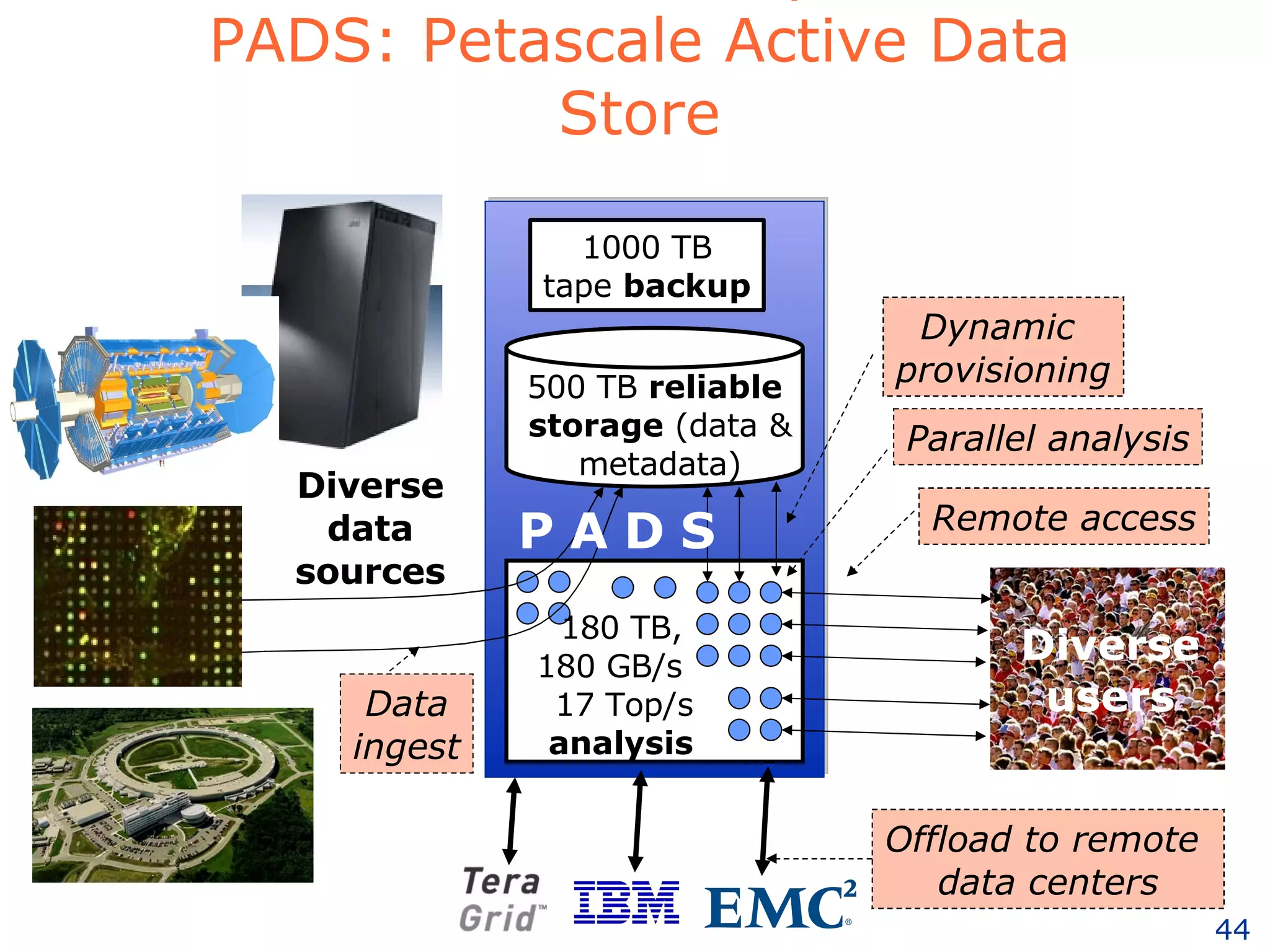
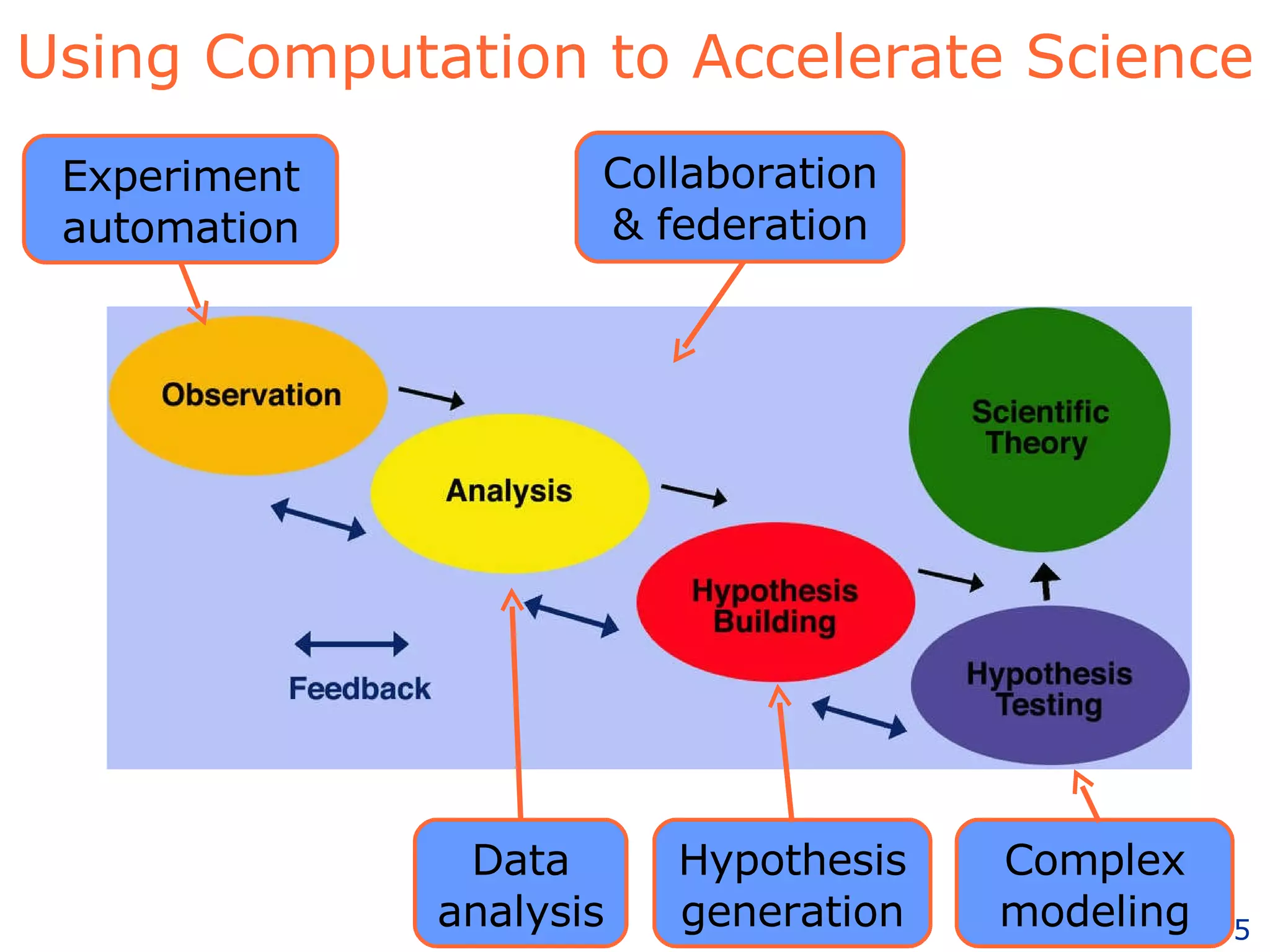
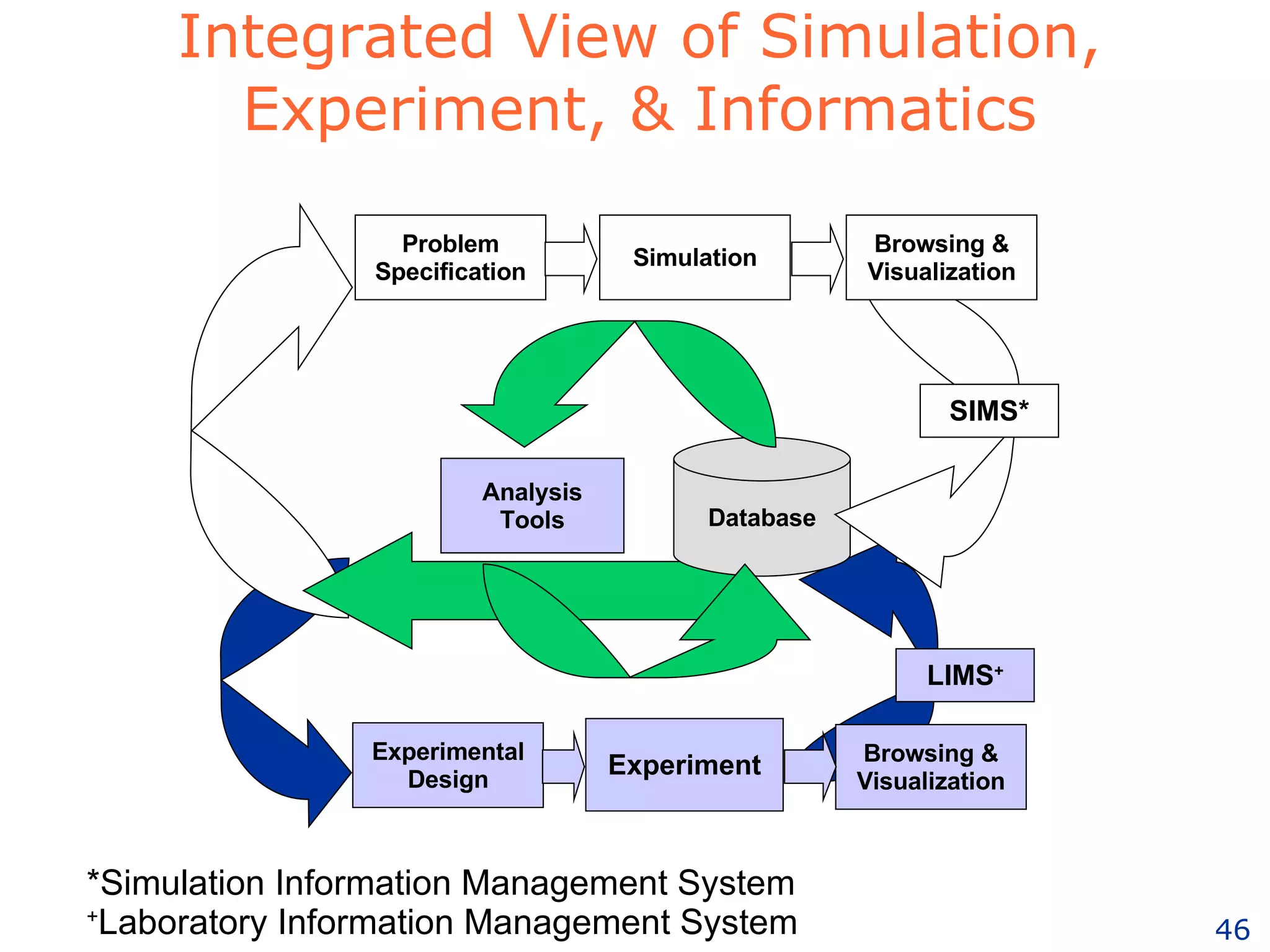
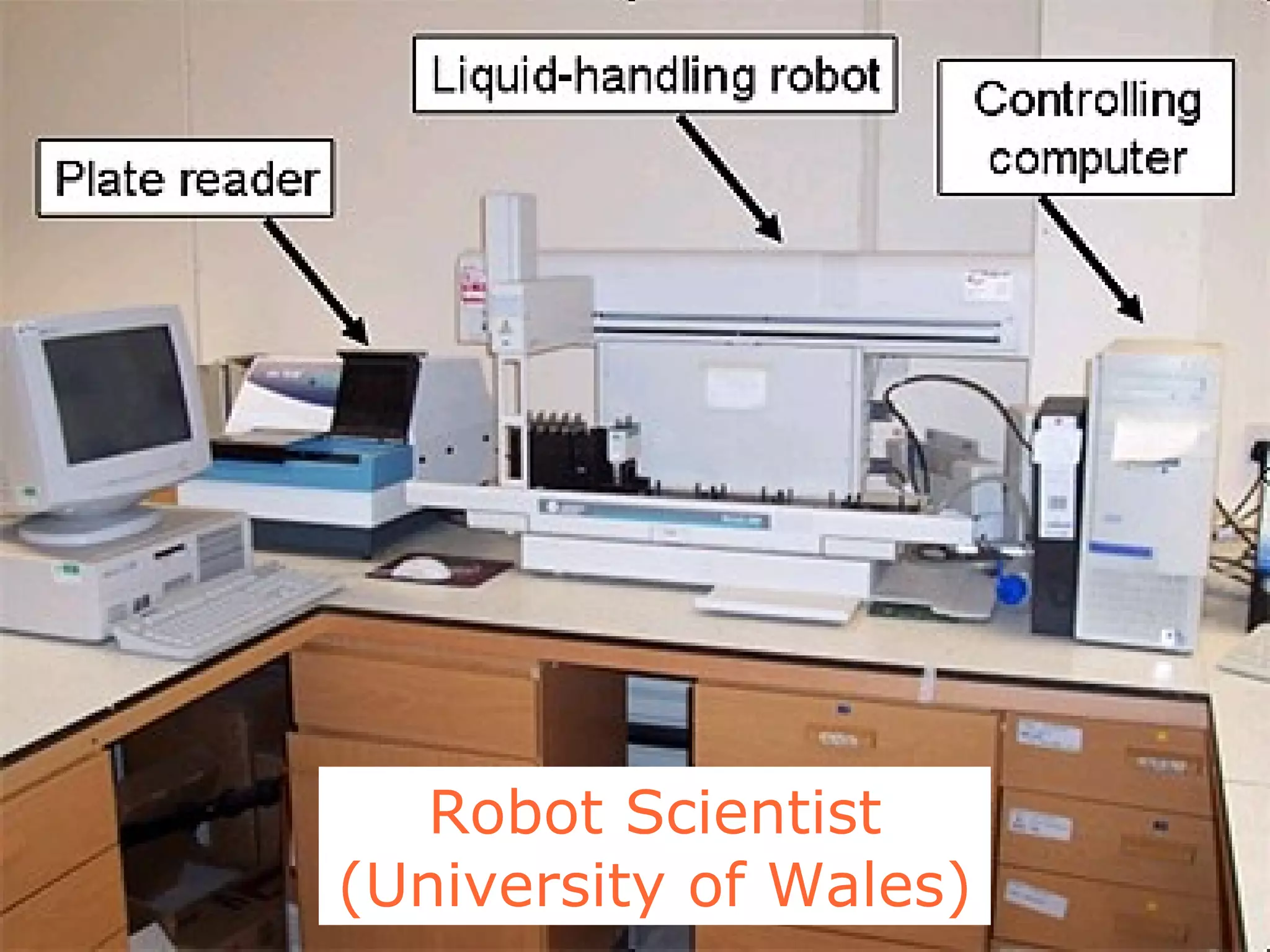
The document discusses how computation can accelerate the generation of new knowledge by enabling large-scale collaborative research and extracting insights from vast amounts of data. It provides examples from astronomy, physics simulations, and biomedical research where computation has allowed more data and researchers to be incorporated, advancing various fields more quickly over time. Computation allows for data sharing, analysis, and hypothesis generation at scales not previously possible.