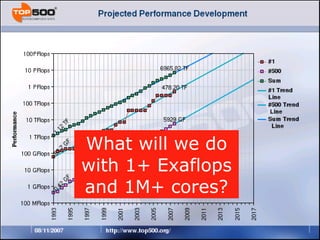
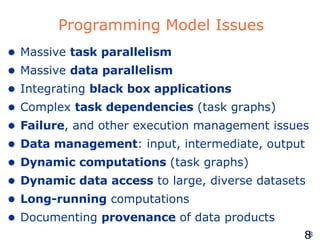
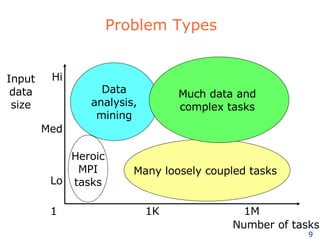
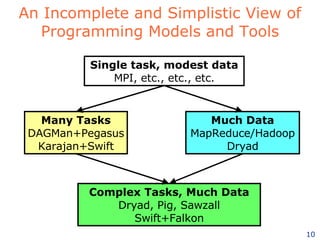
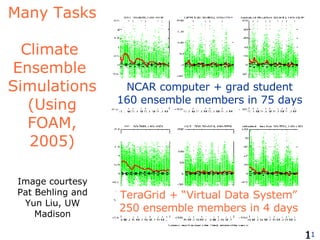
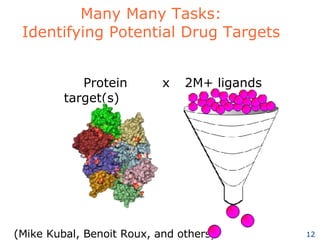
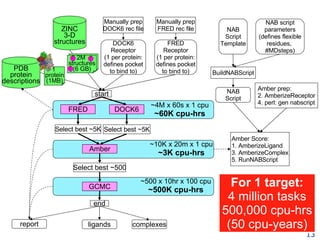
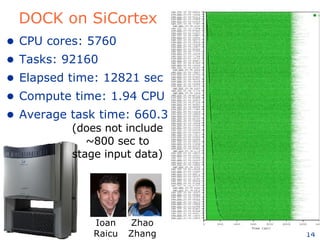
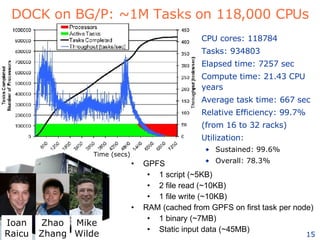
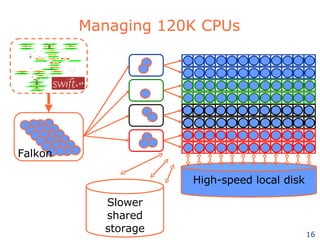
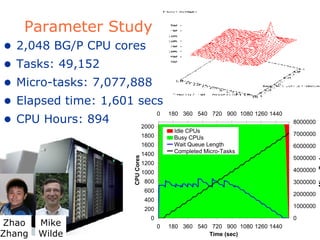
The document discusses how new supercomputing applications are increasingly focused on "logistical" issues like executing many communication-intensive tasks over large shared datasets, rather than "heroic" computations of a single task. It argues that new programming models and tools are needed to efficiently manage large numbers of tasks, complex data dependencies, and failures at extreme scales of petascale and exascale computers. Examples of applications that could benefit include parameter studies, ensemble simulations, data analysis, and scientific workflows involving millions of tasks.