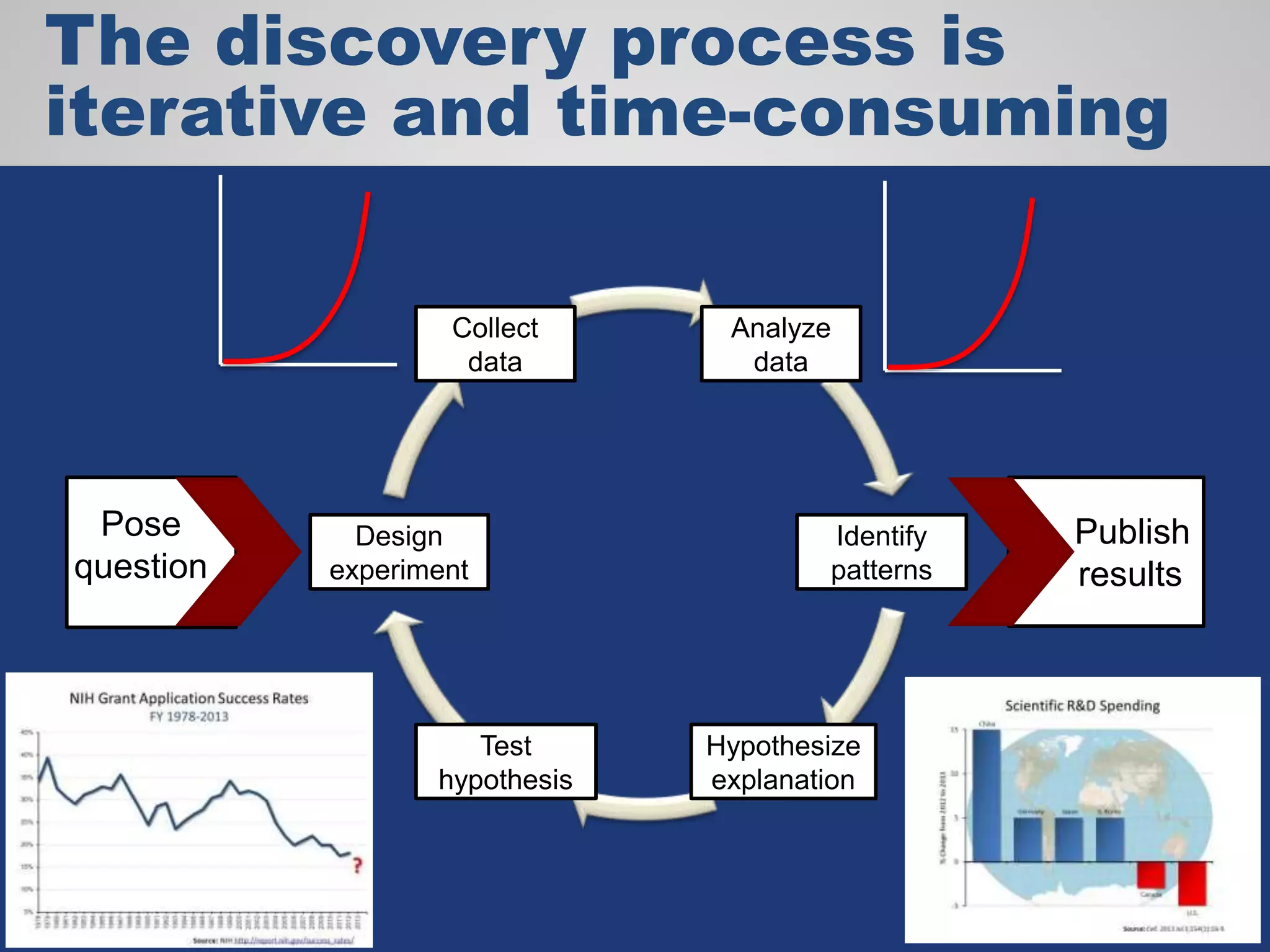
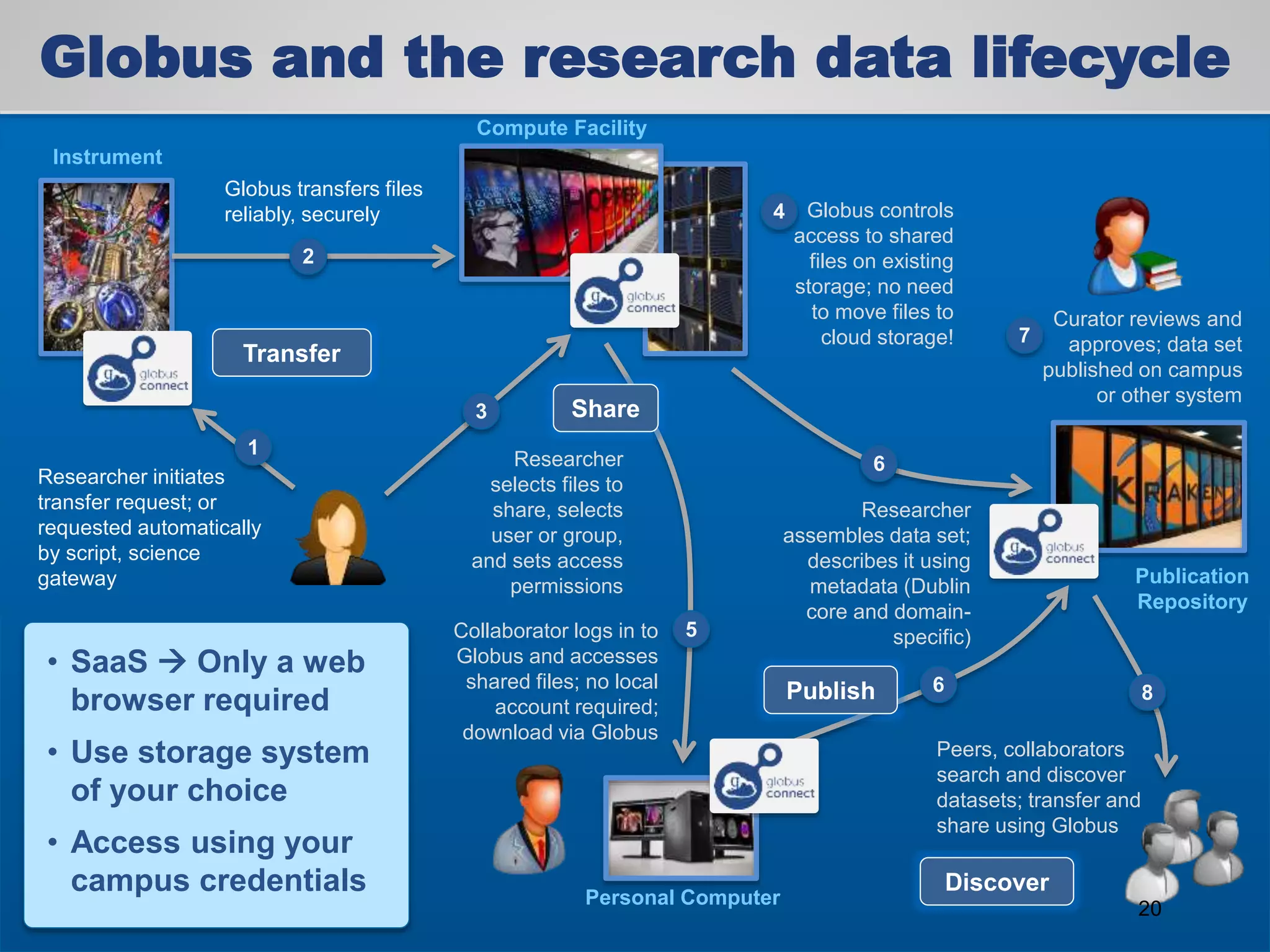
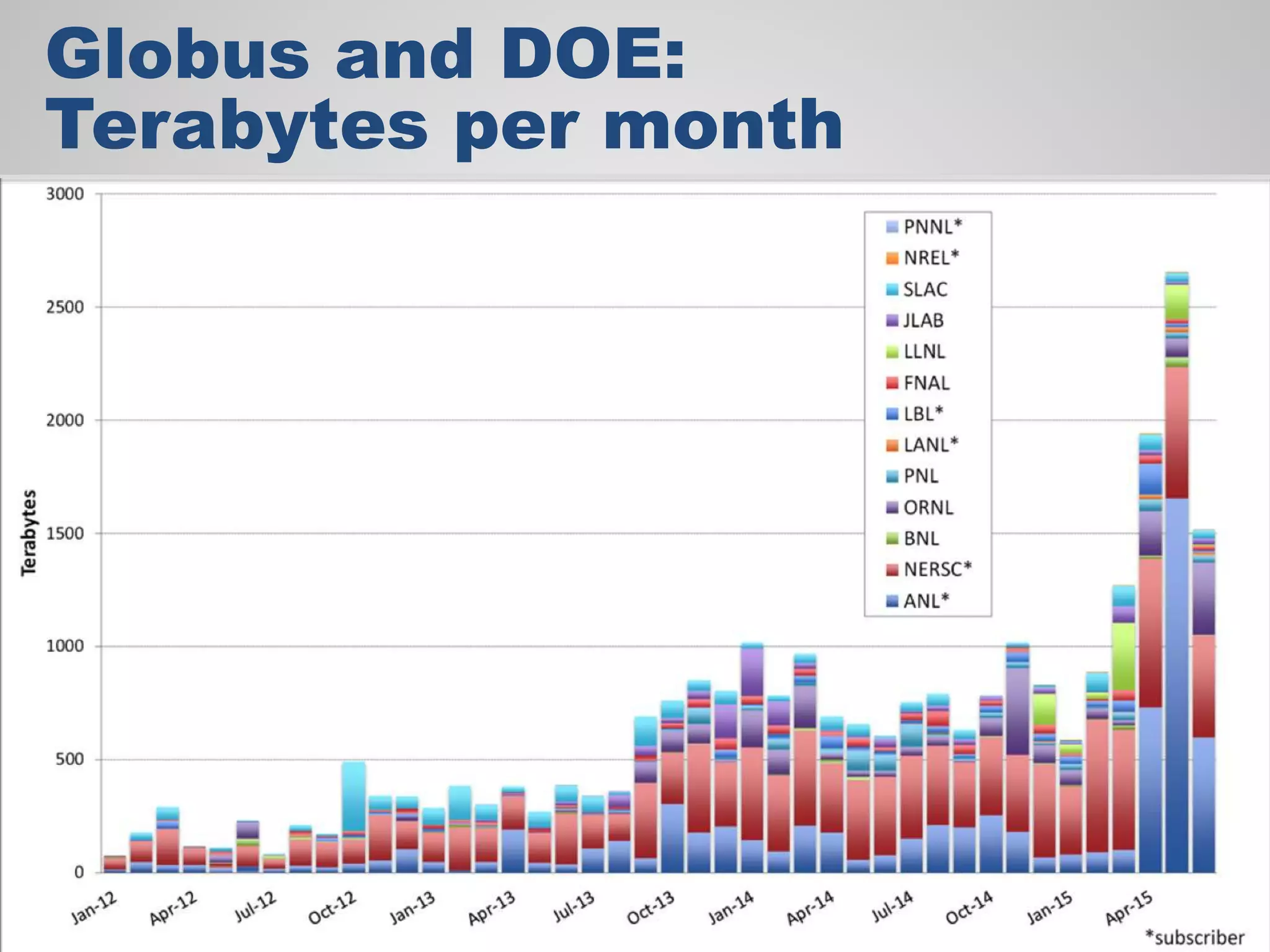
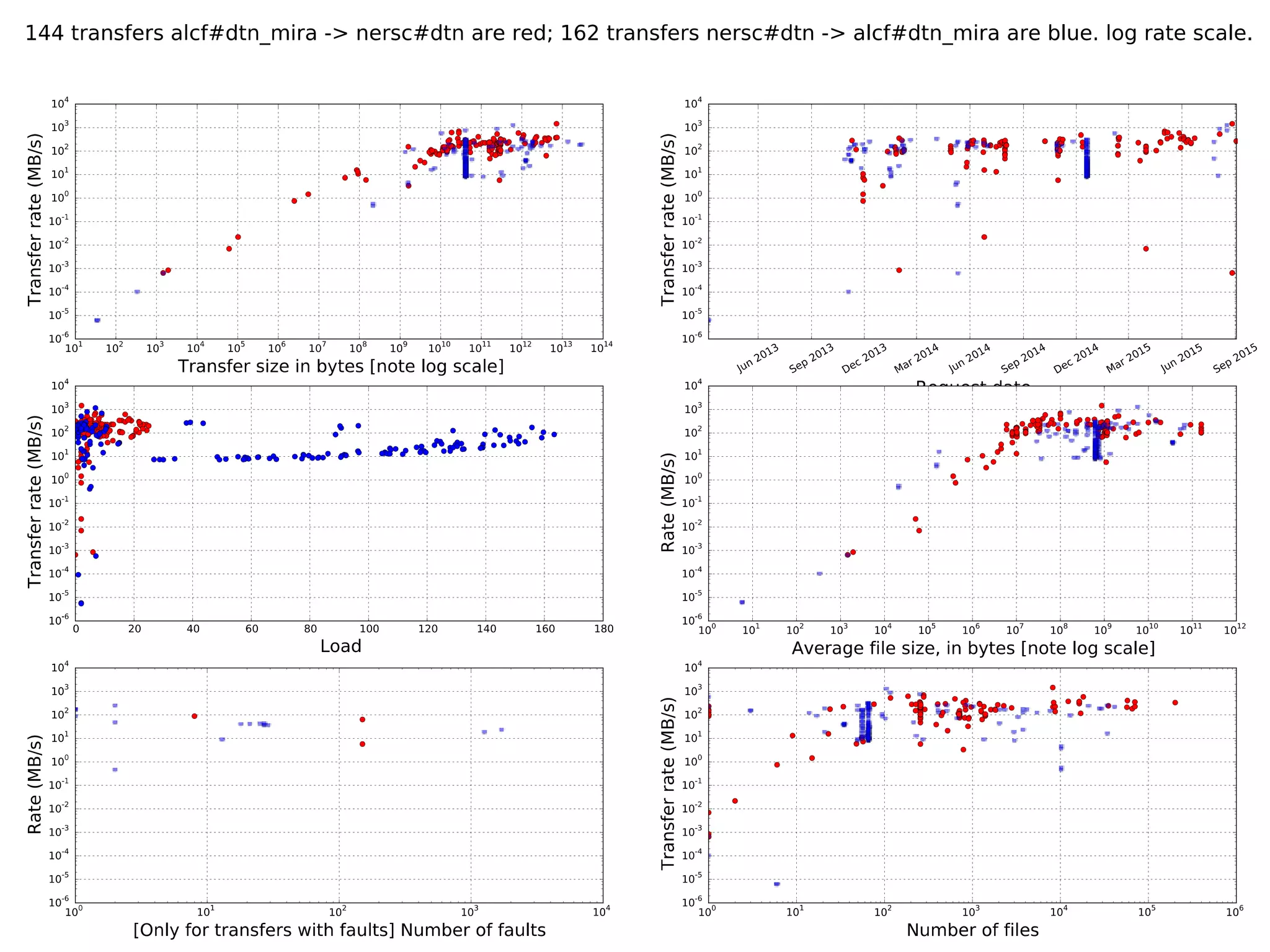
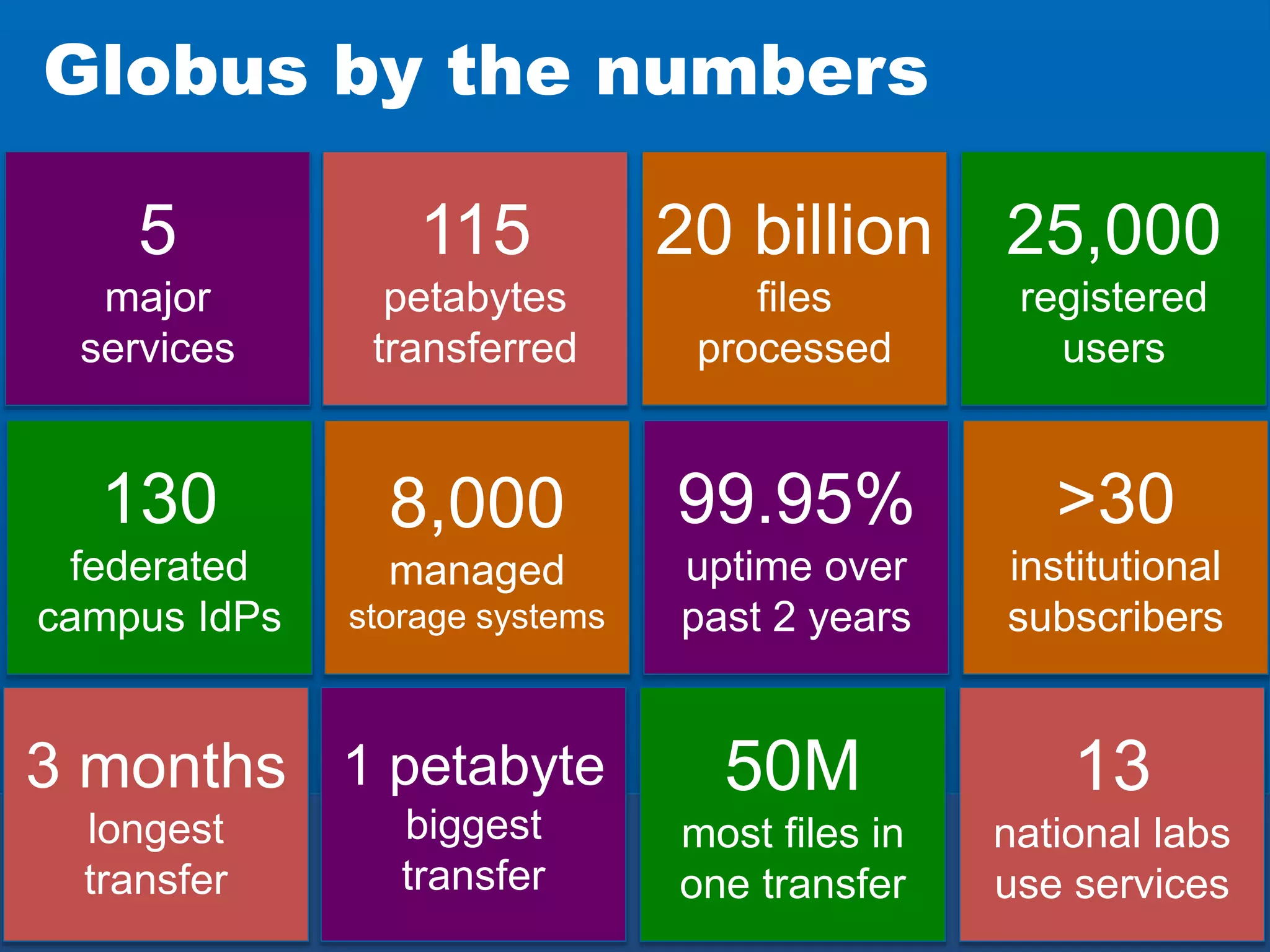
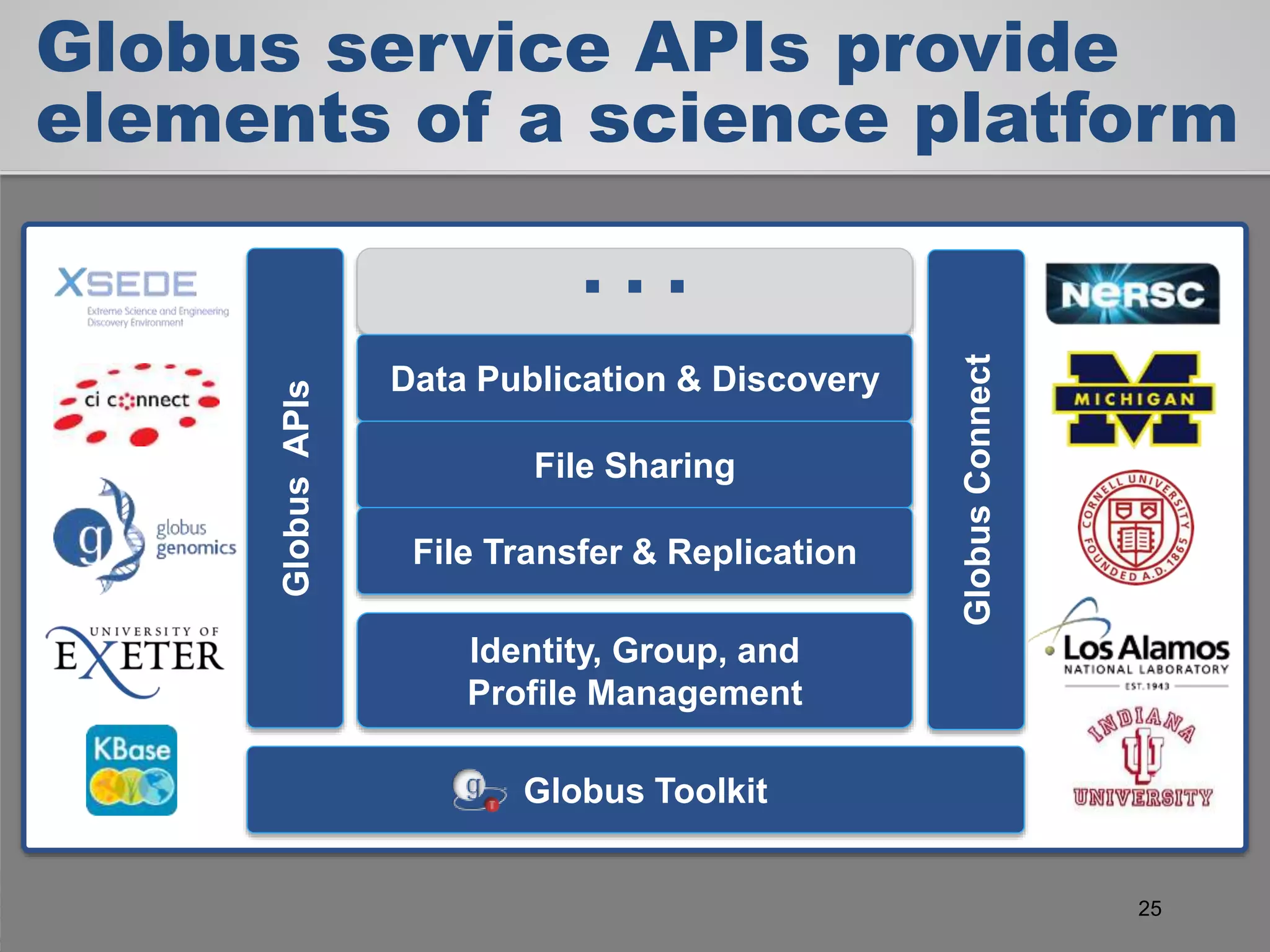
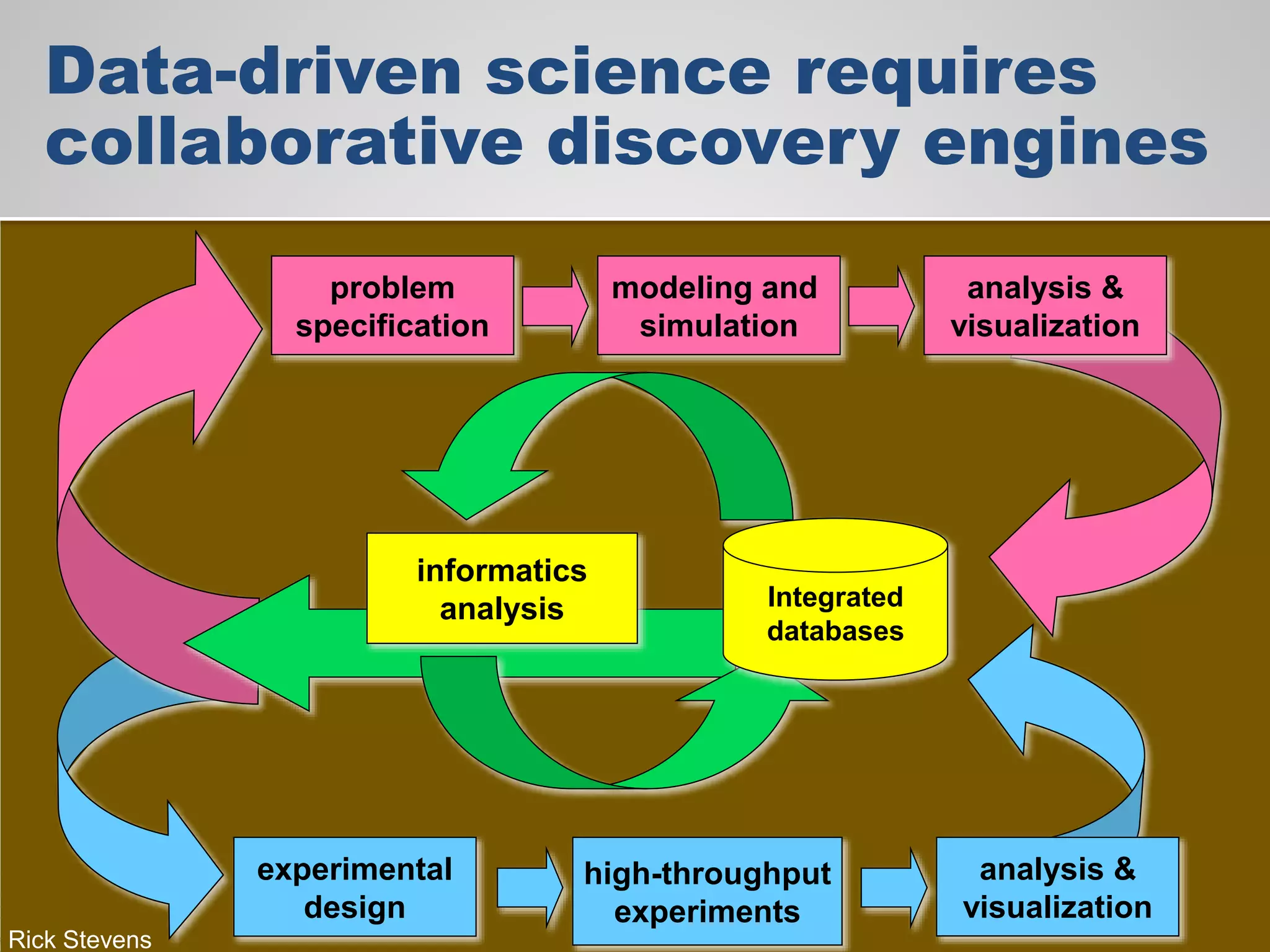
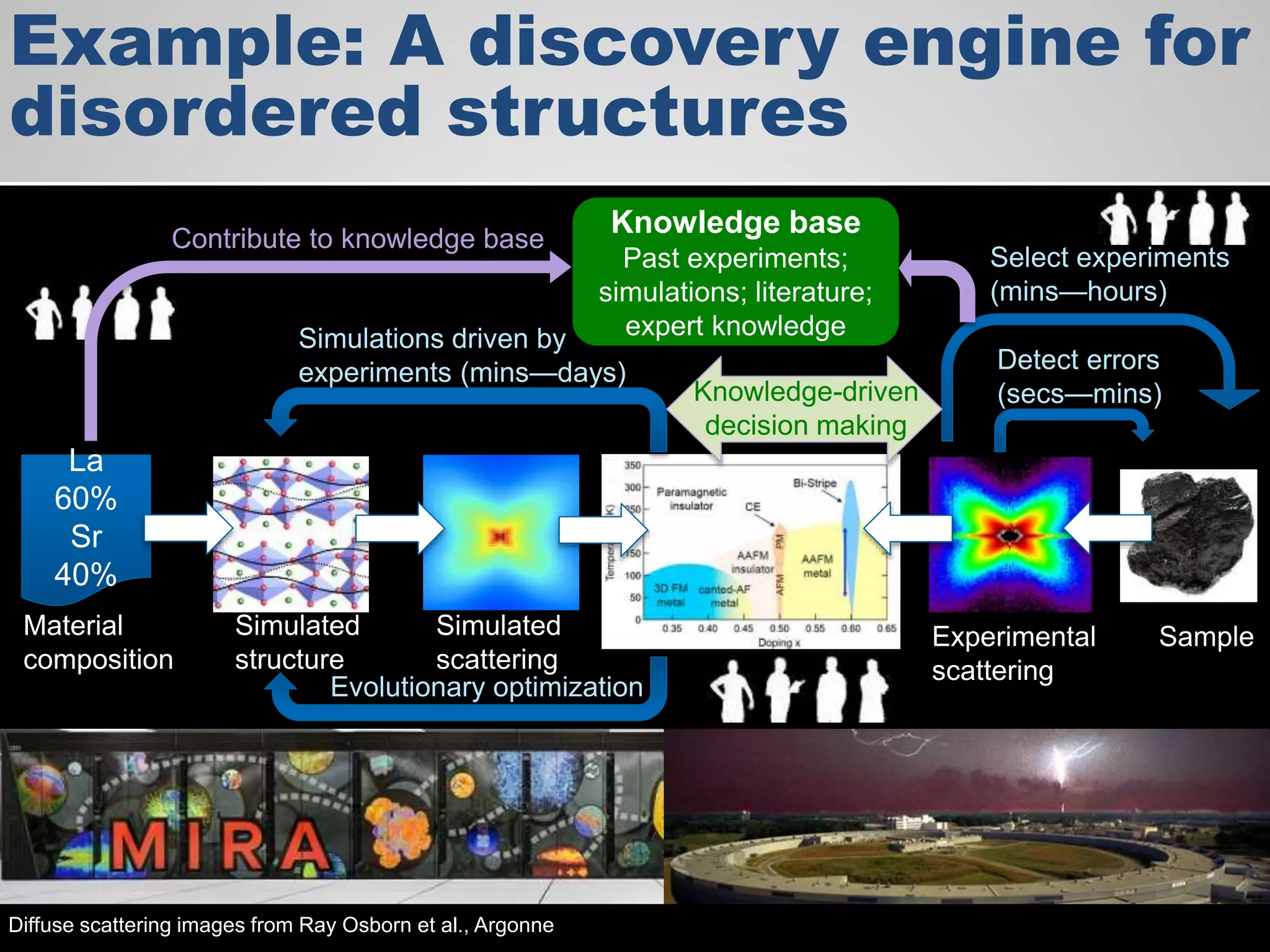
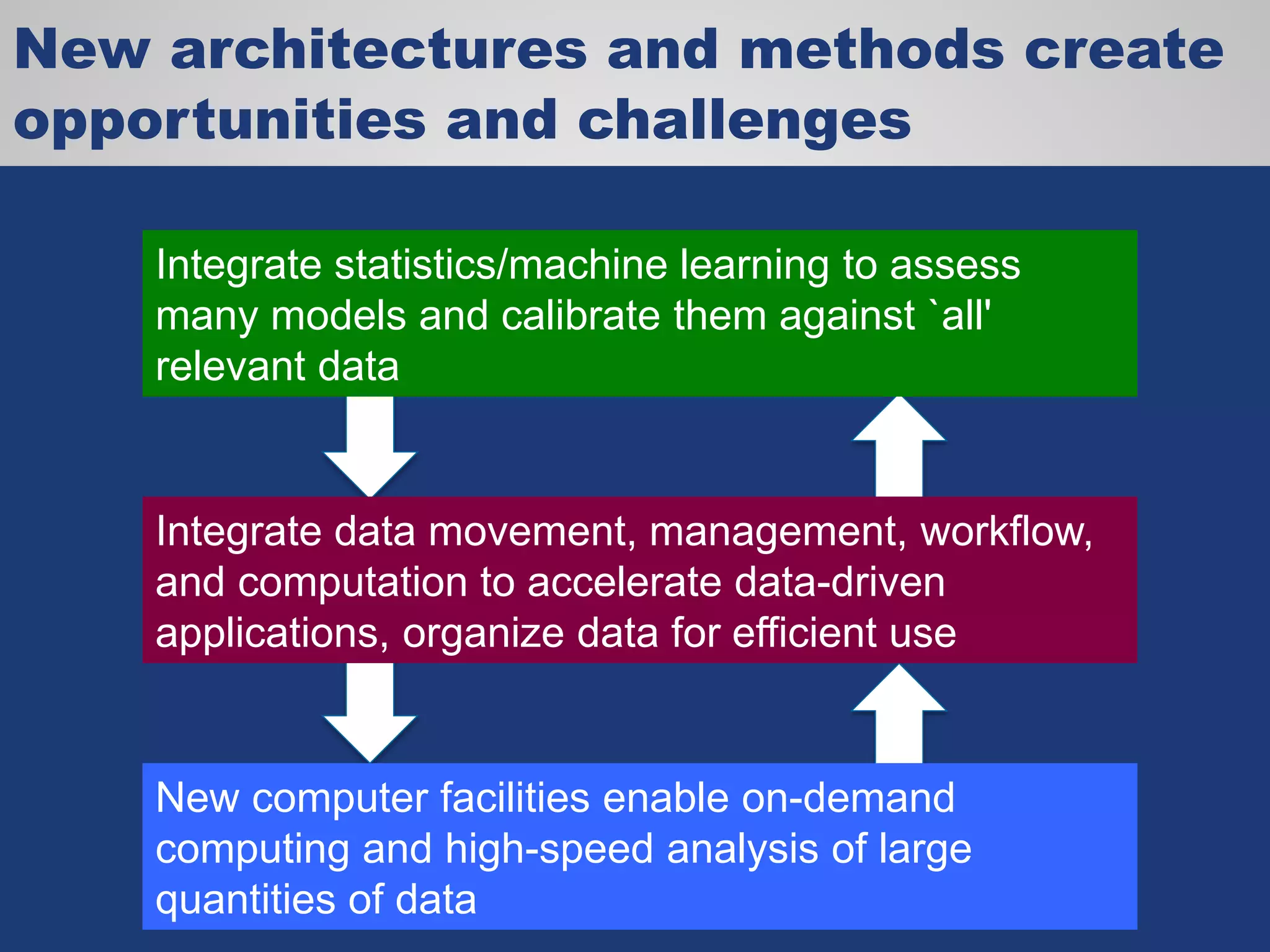
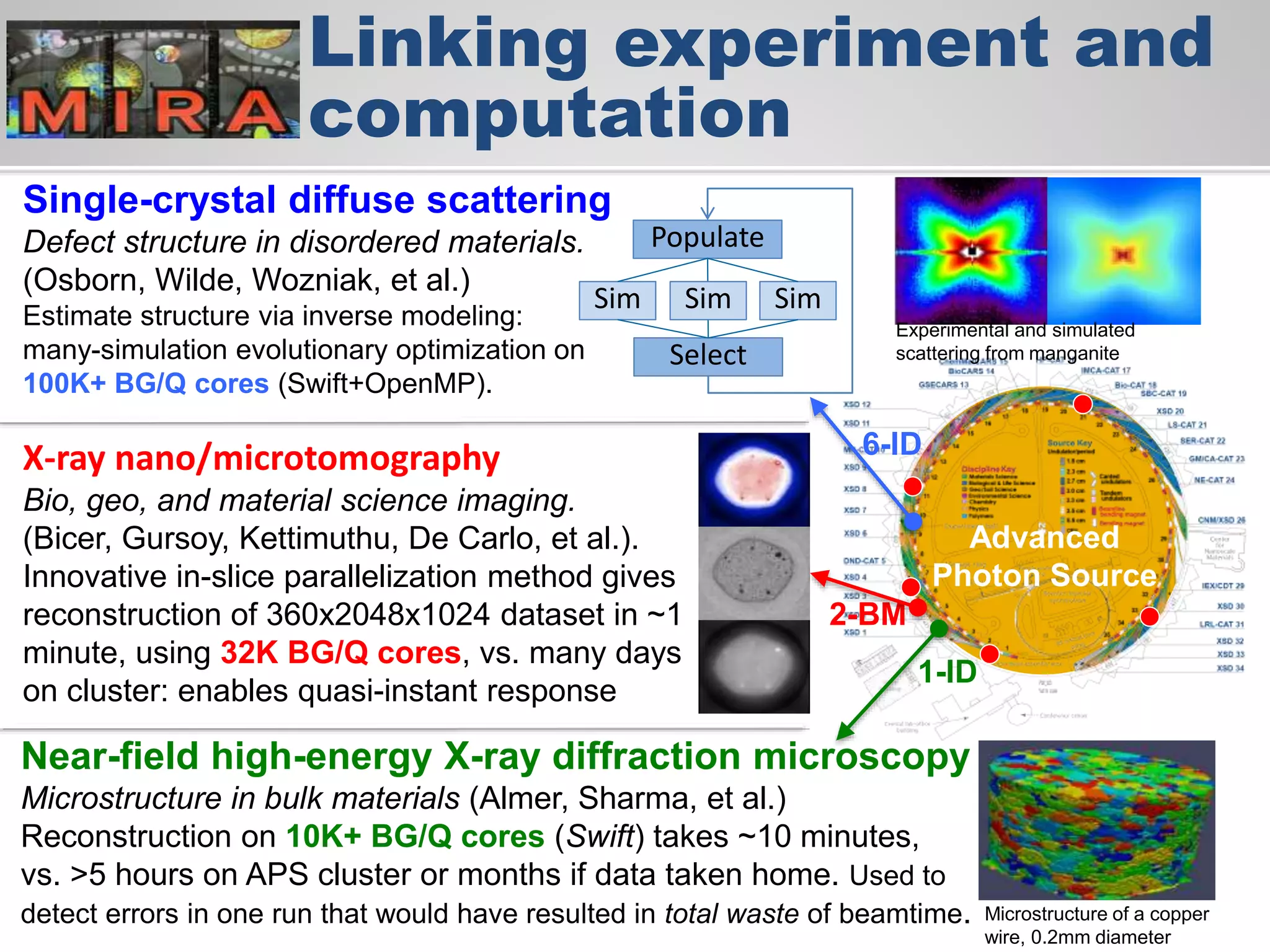
The document discusses the acceleration of scientific discovery through the use of automated data management services, outsourcing, and cloud-based platforms like Globus. It highlights the importance of eliminating data friction and creating discovery engines to enhance collaboration and efficiency in scientific research. Key themes include the automation of data publication, sharing, and analysis, as well as the need for services that address the entire research lifecycle.