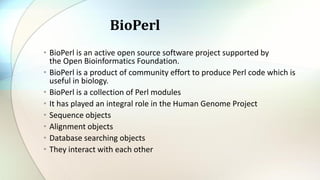
Bioperl is an open-source software project developed for bioinformatics, providing a collection of Perl modules that facilitate biological data handling. Key modules include Bio::Seq and Bio::SeqIO, which allow for the creation and management of sequence objects and reading/writing various file formats. It has been instrumental in initiatives such as the Human Genome Project.