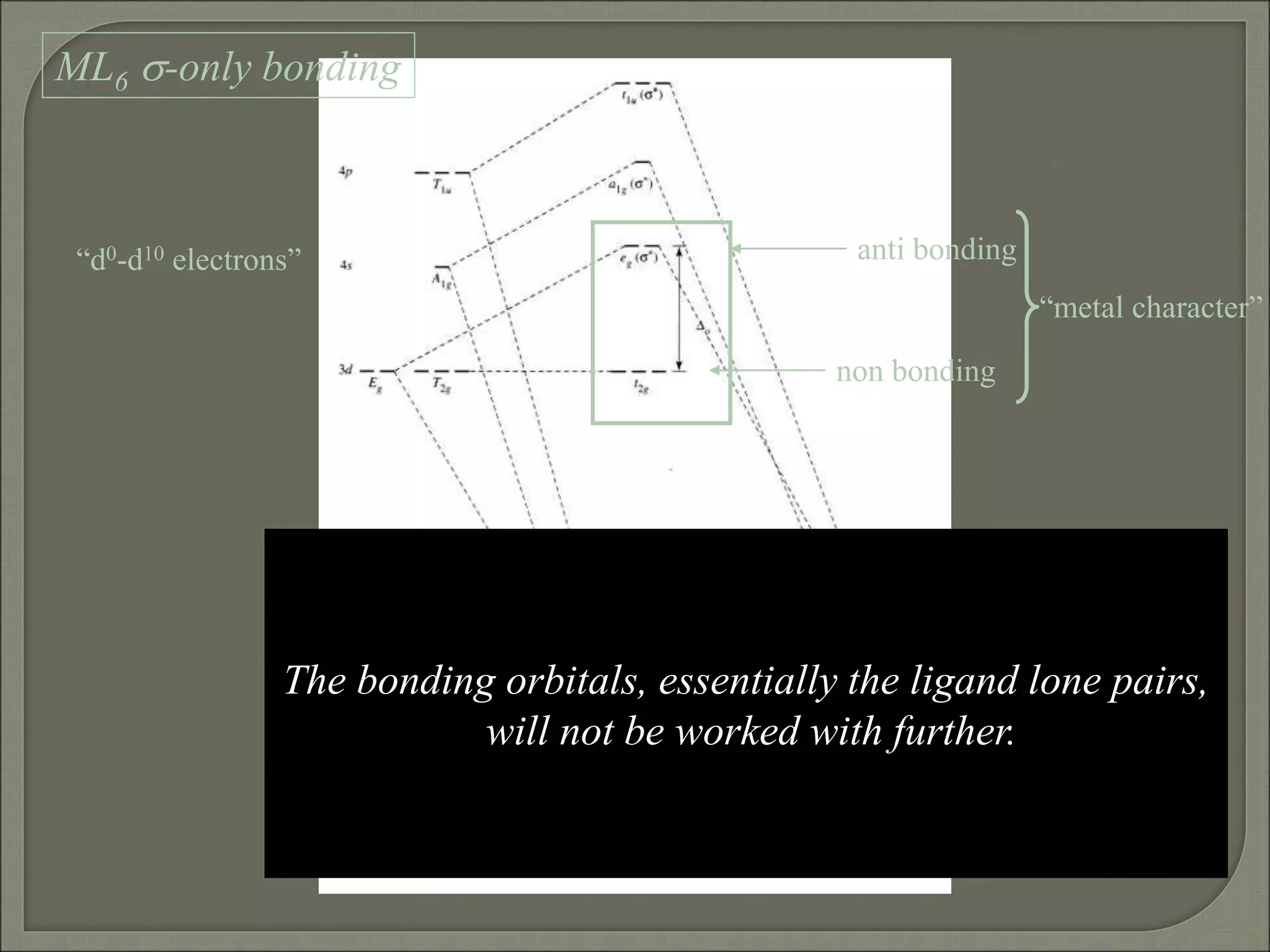
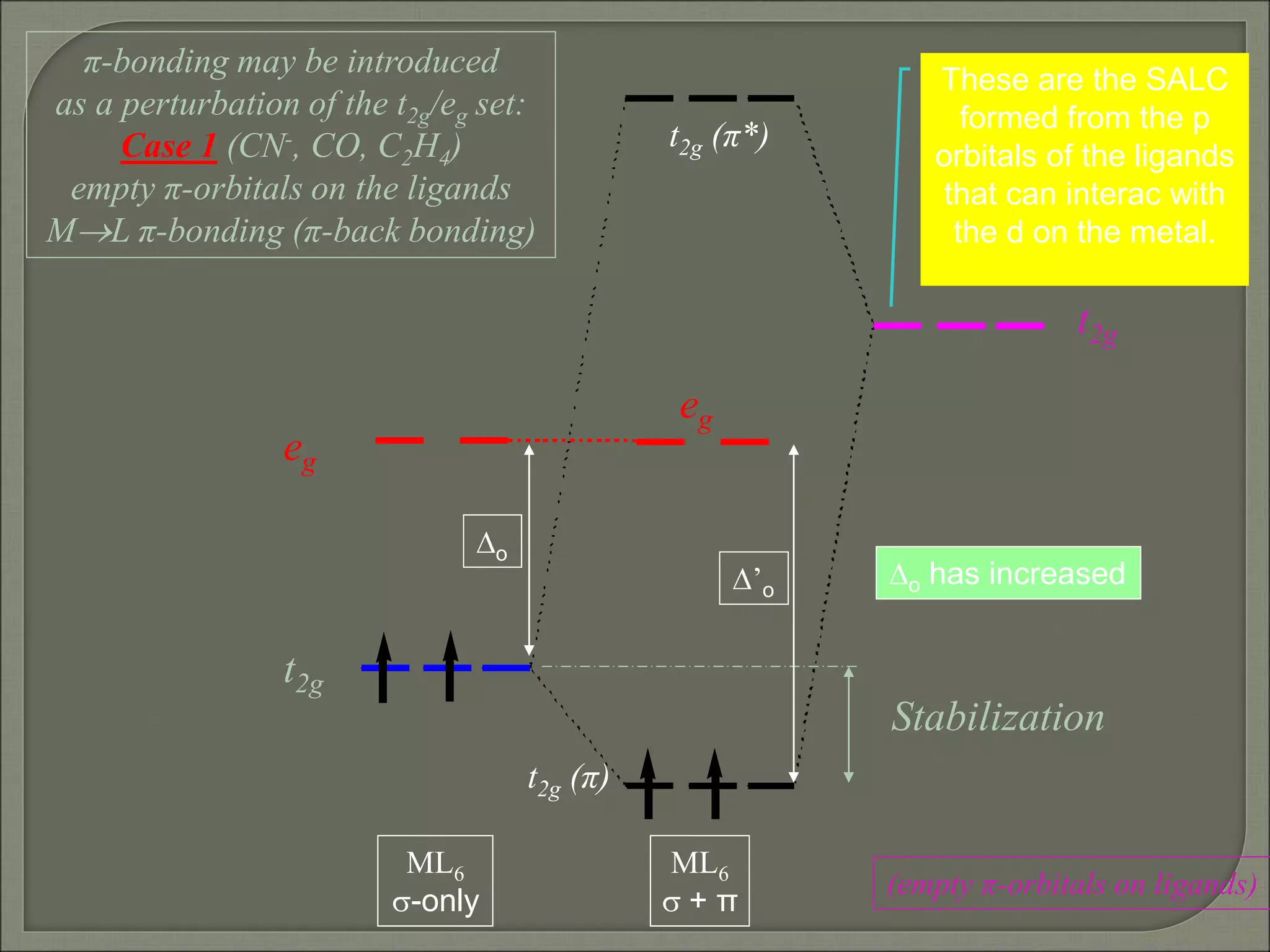
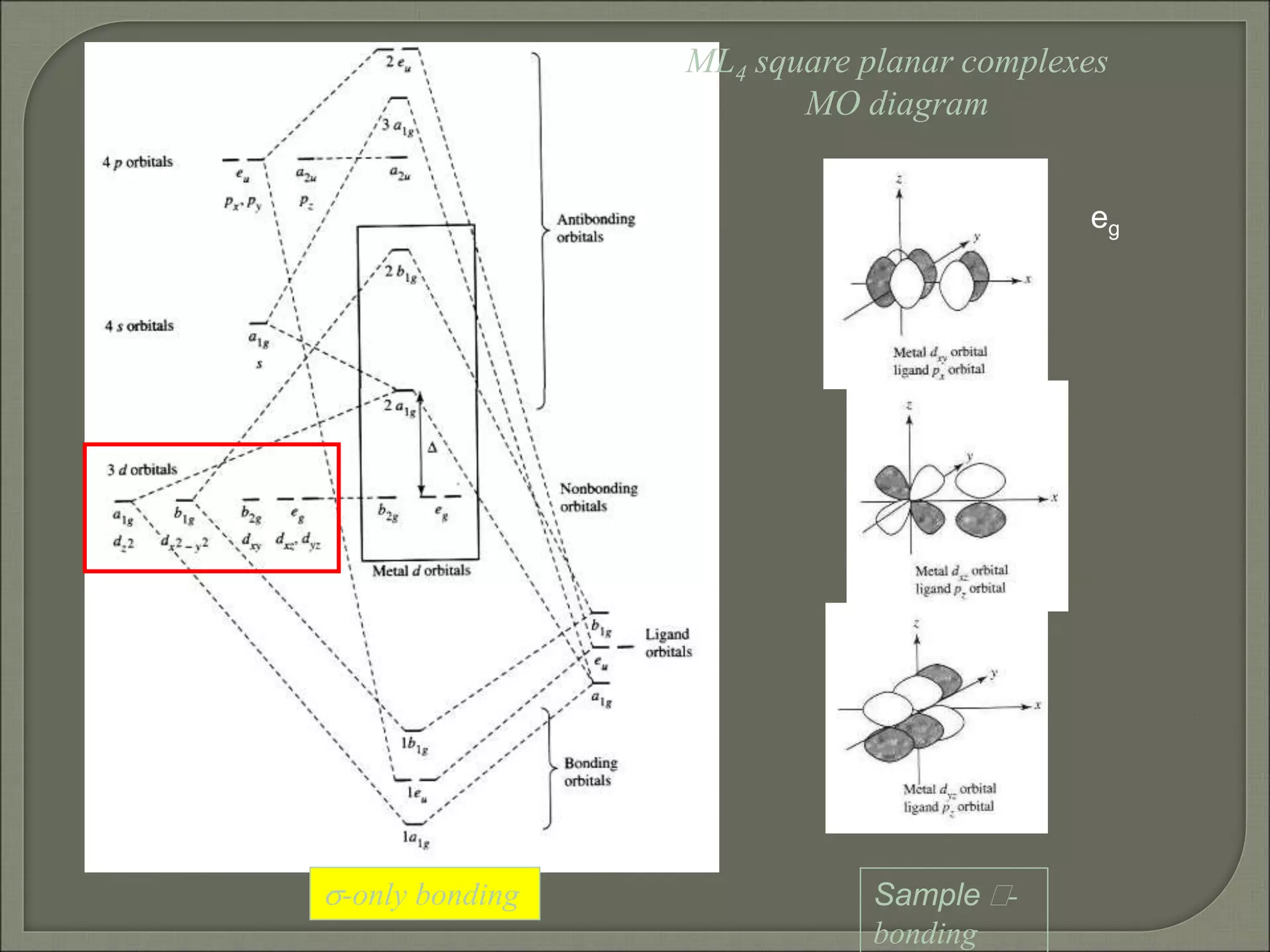
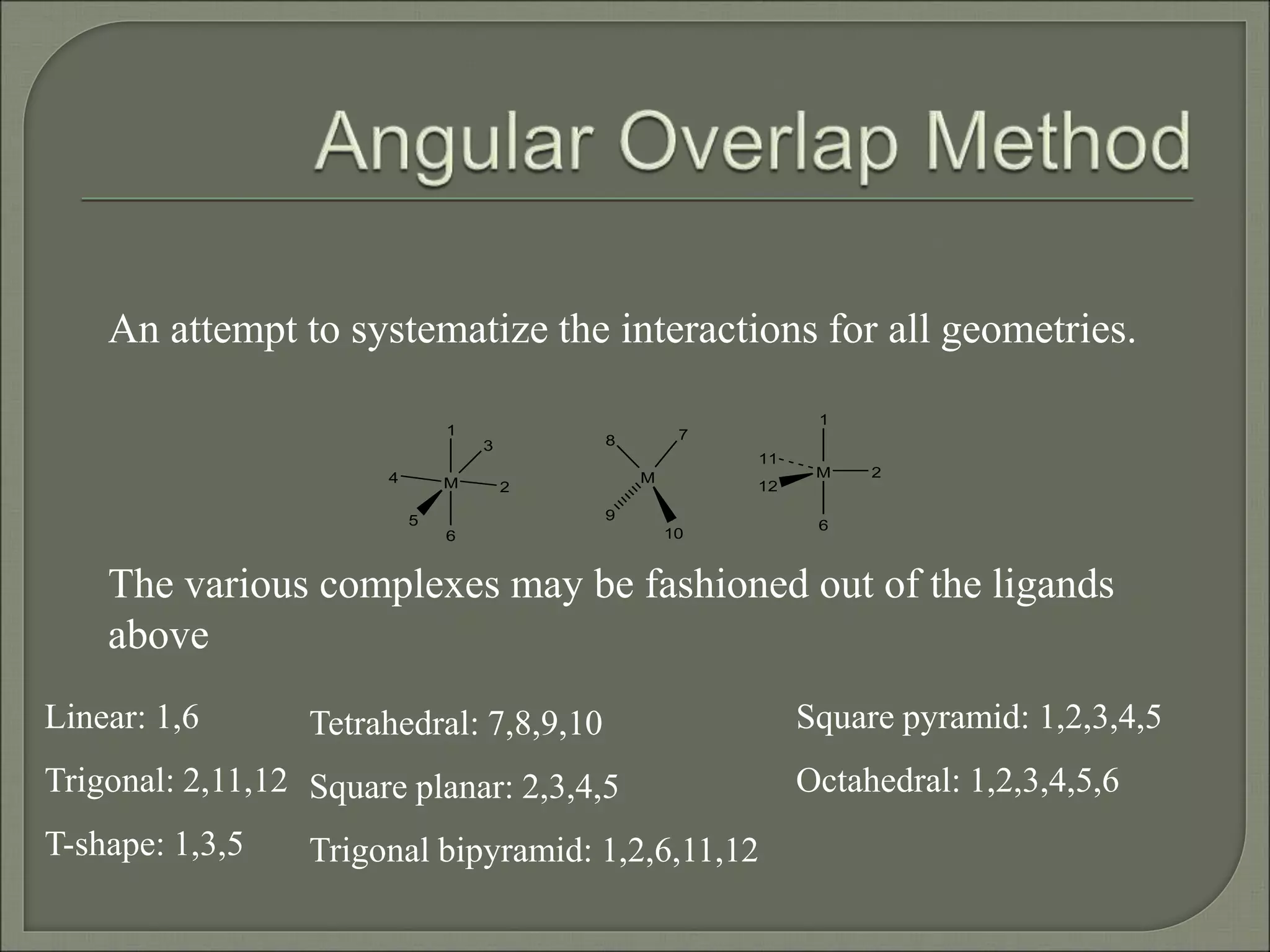
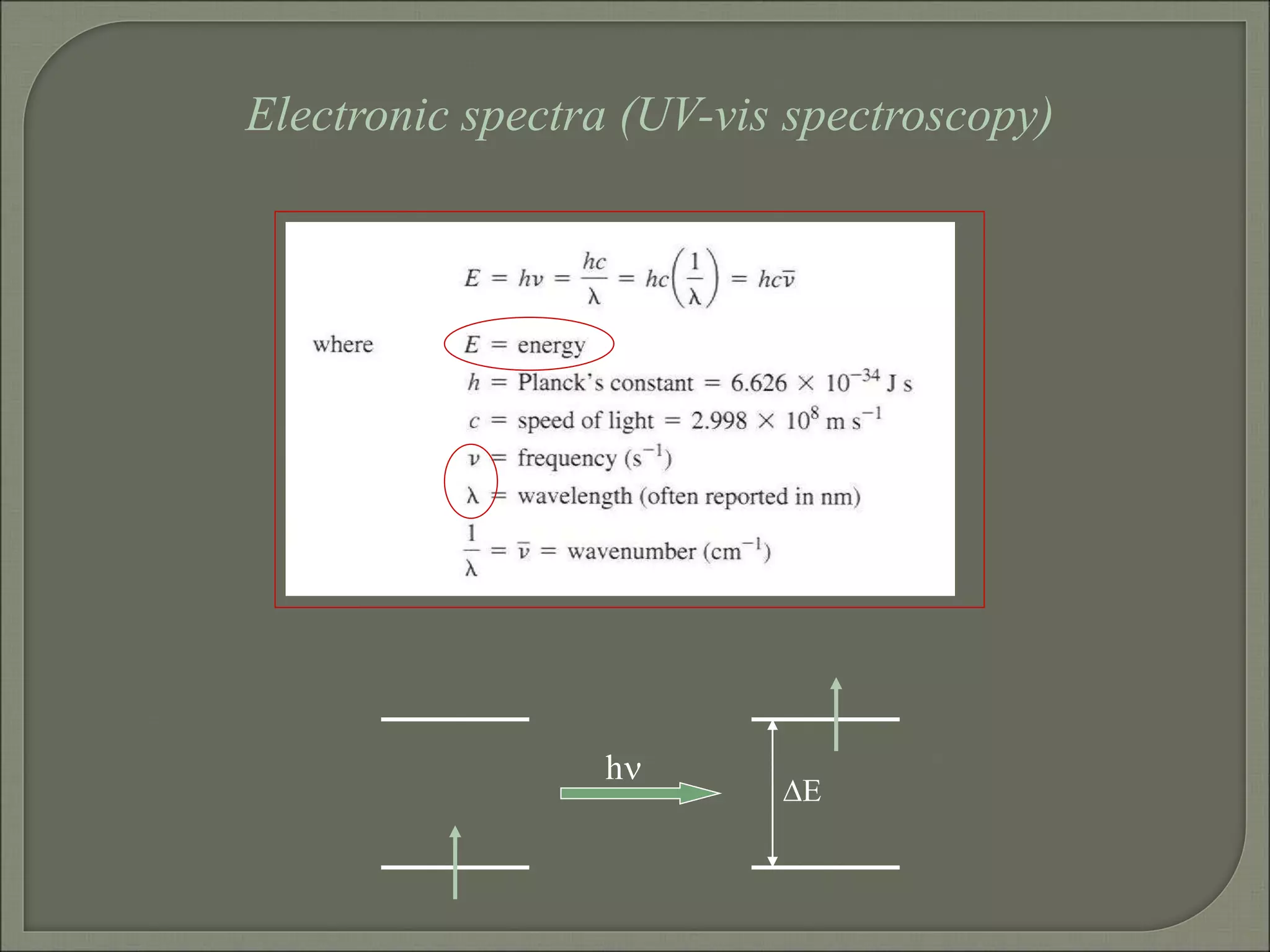
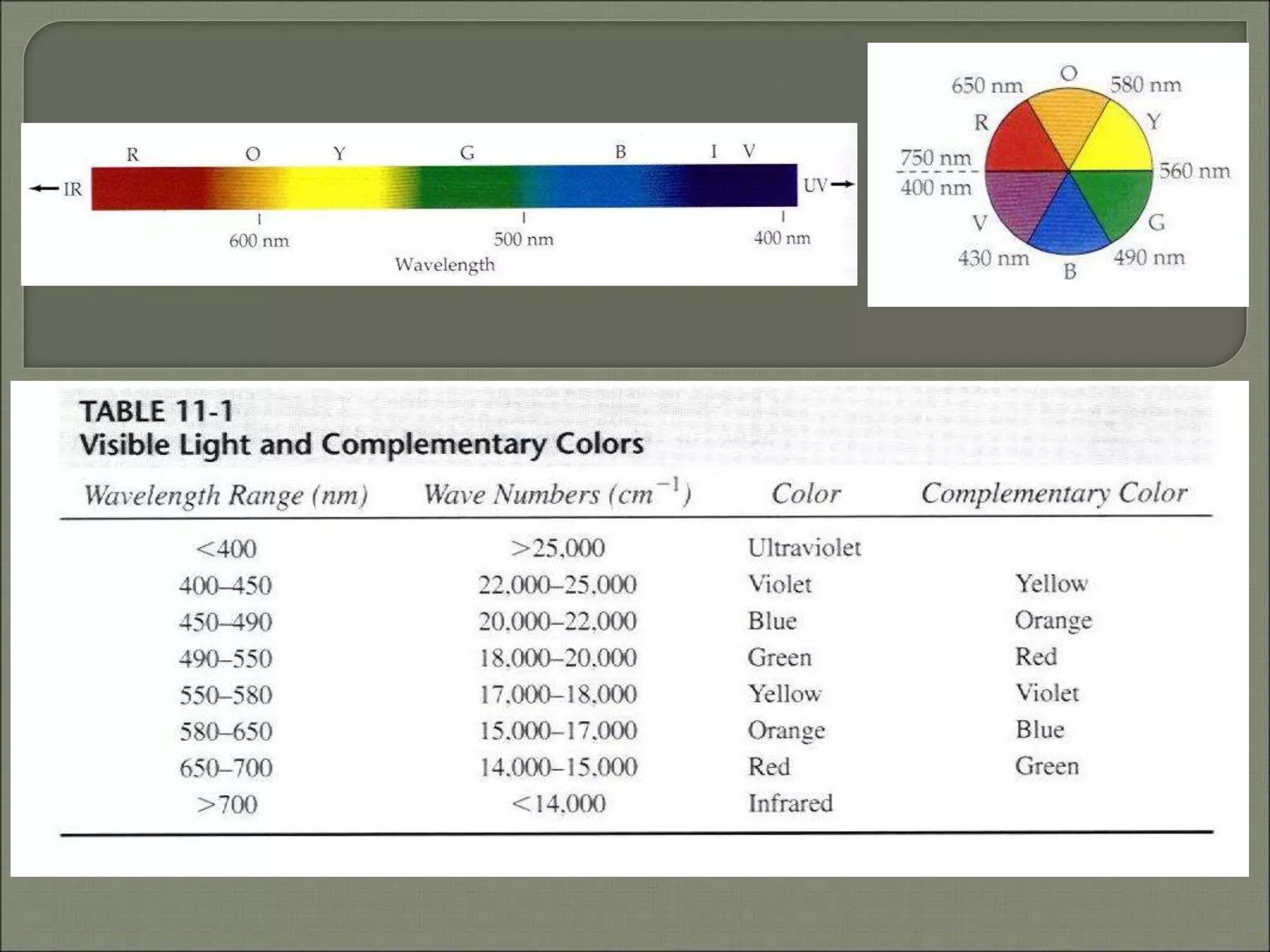
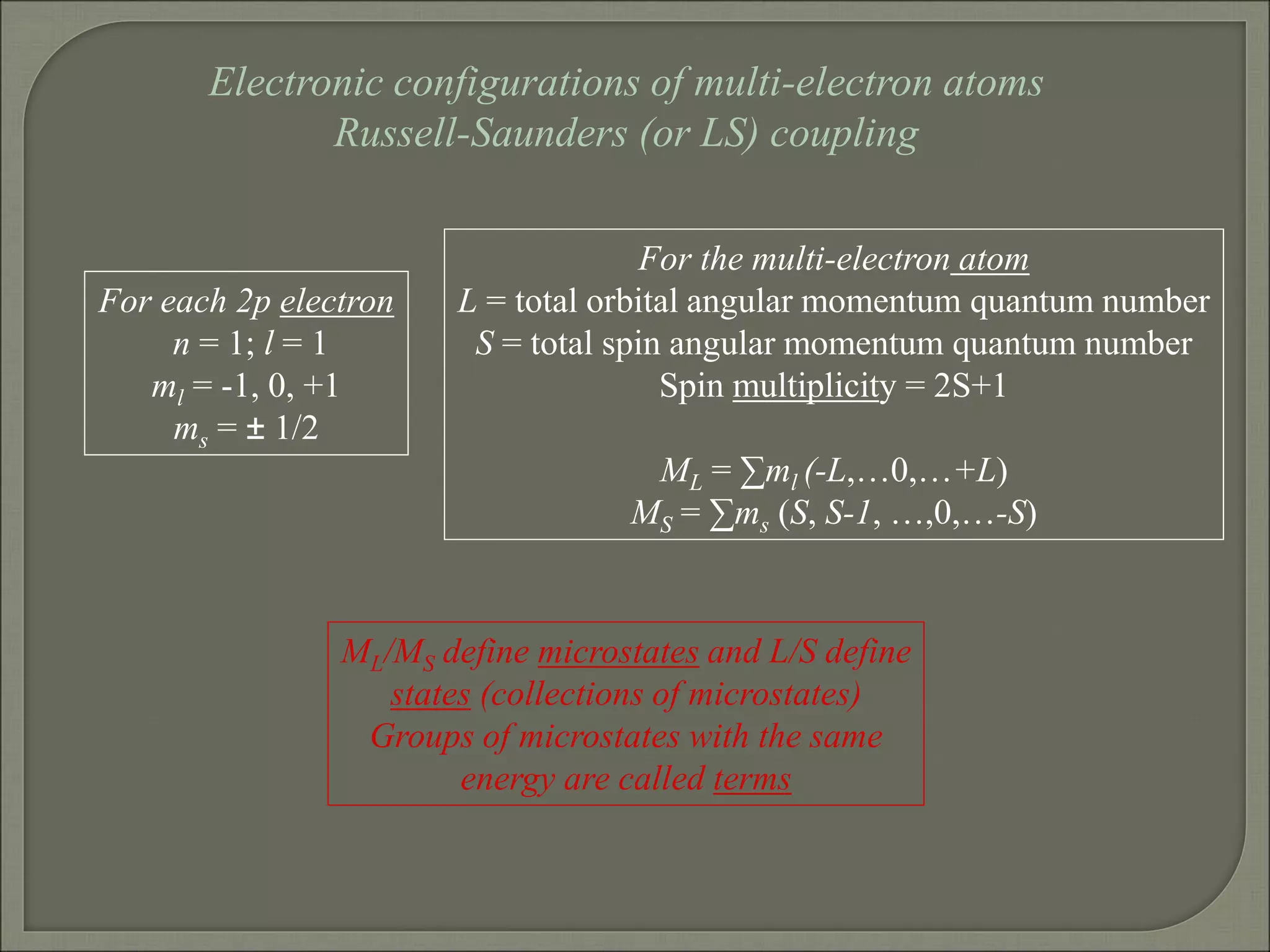
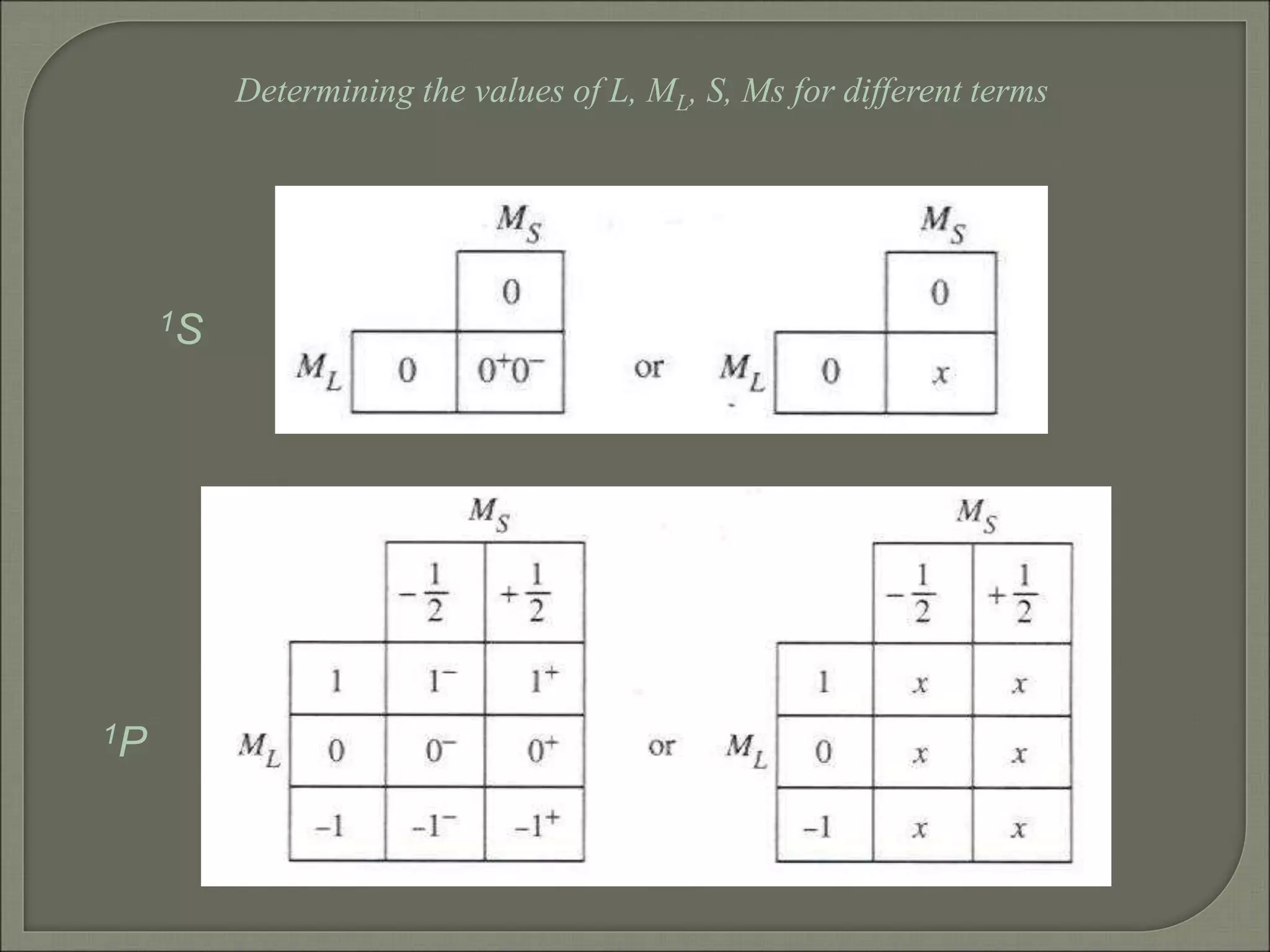
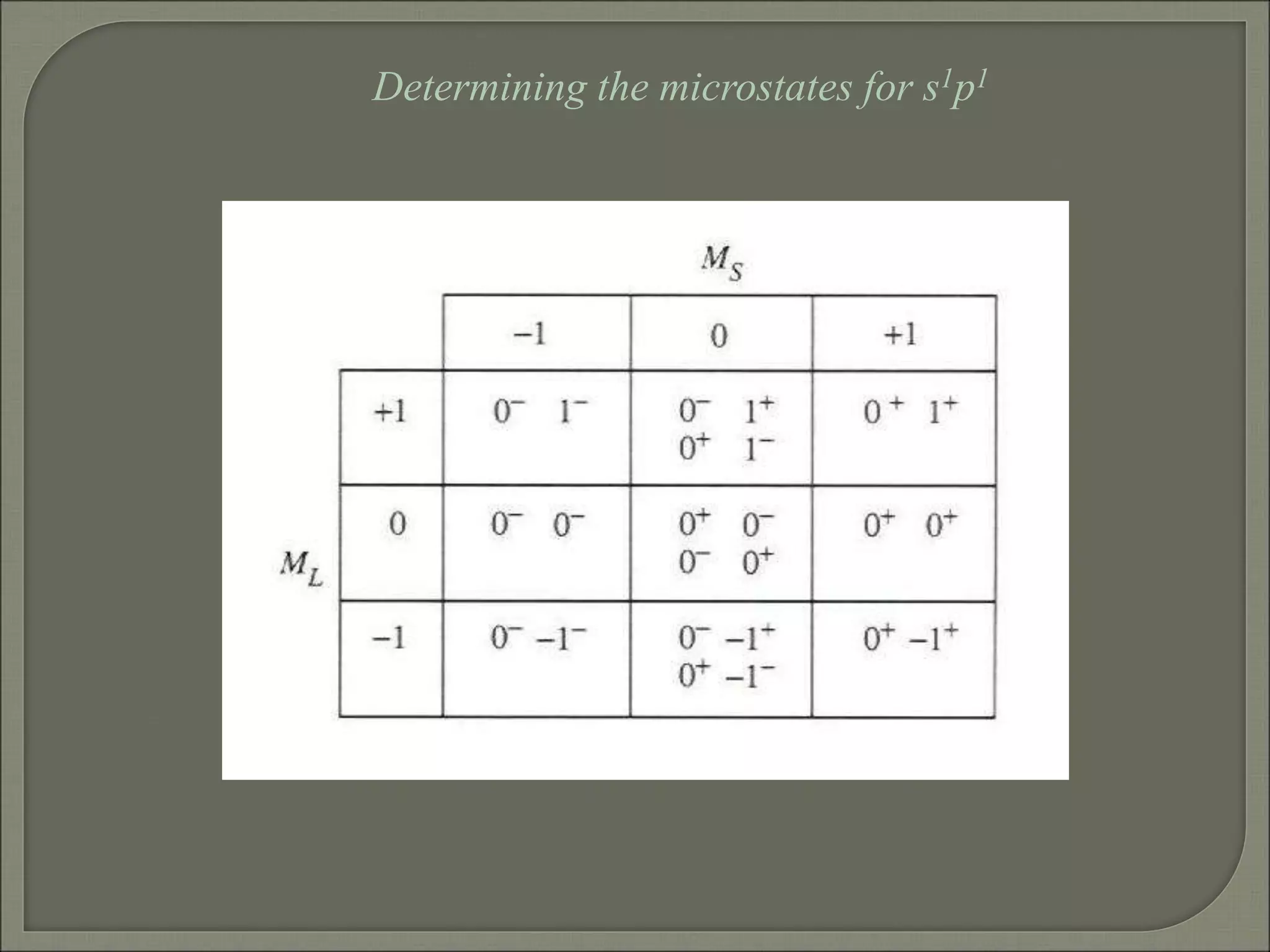
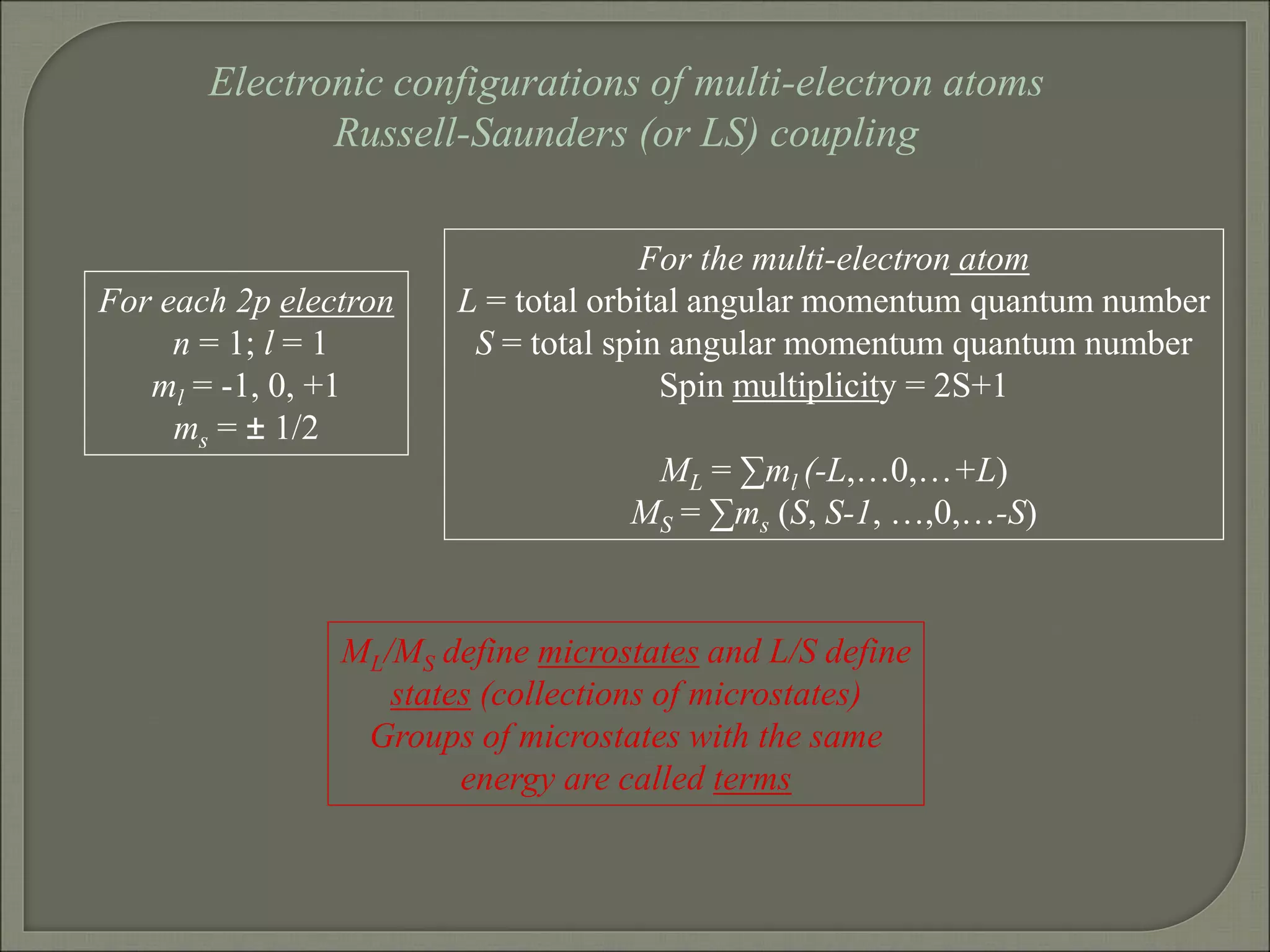
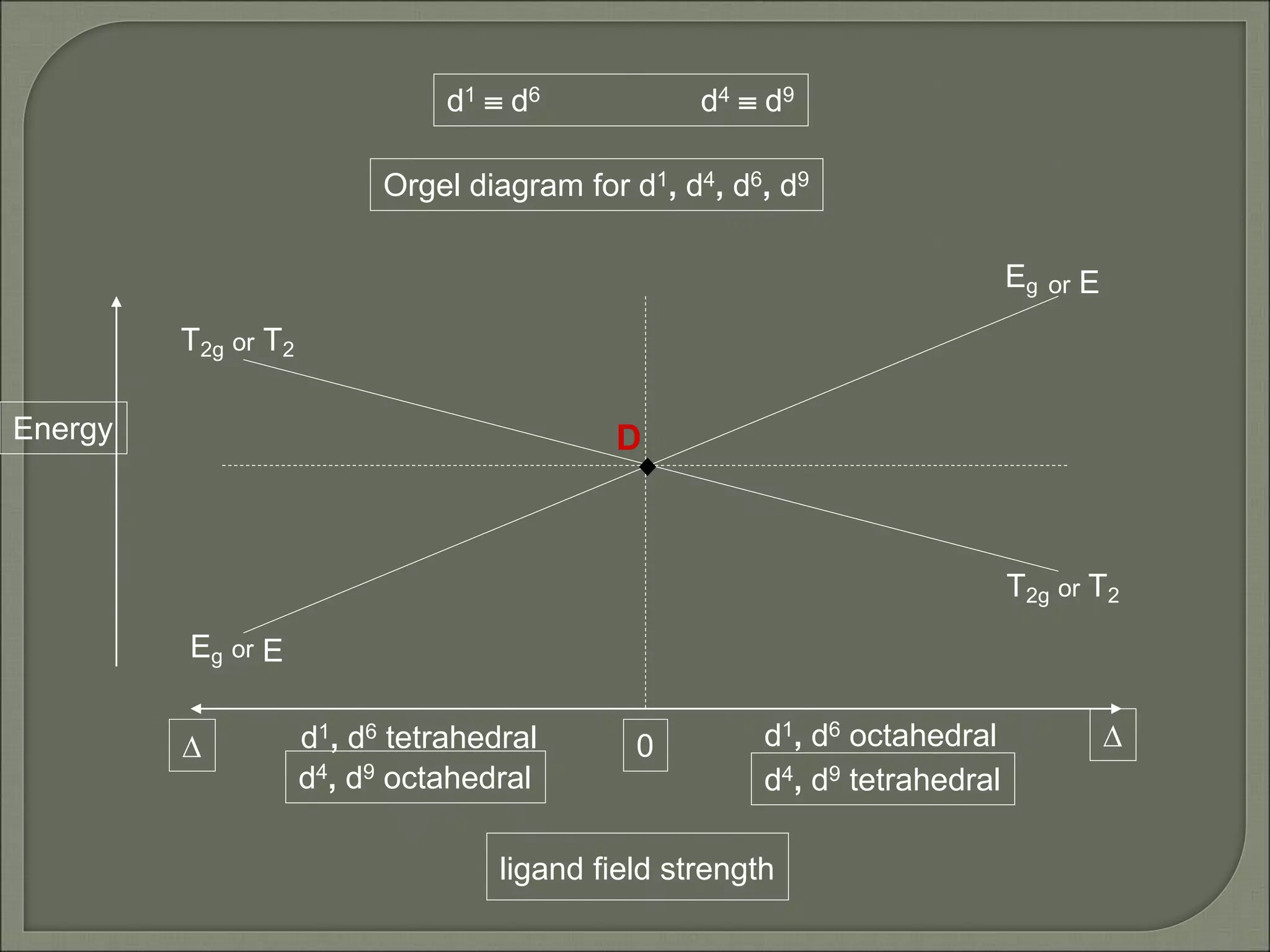
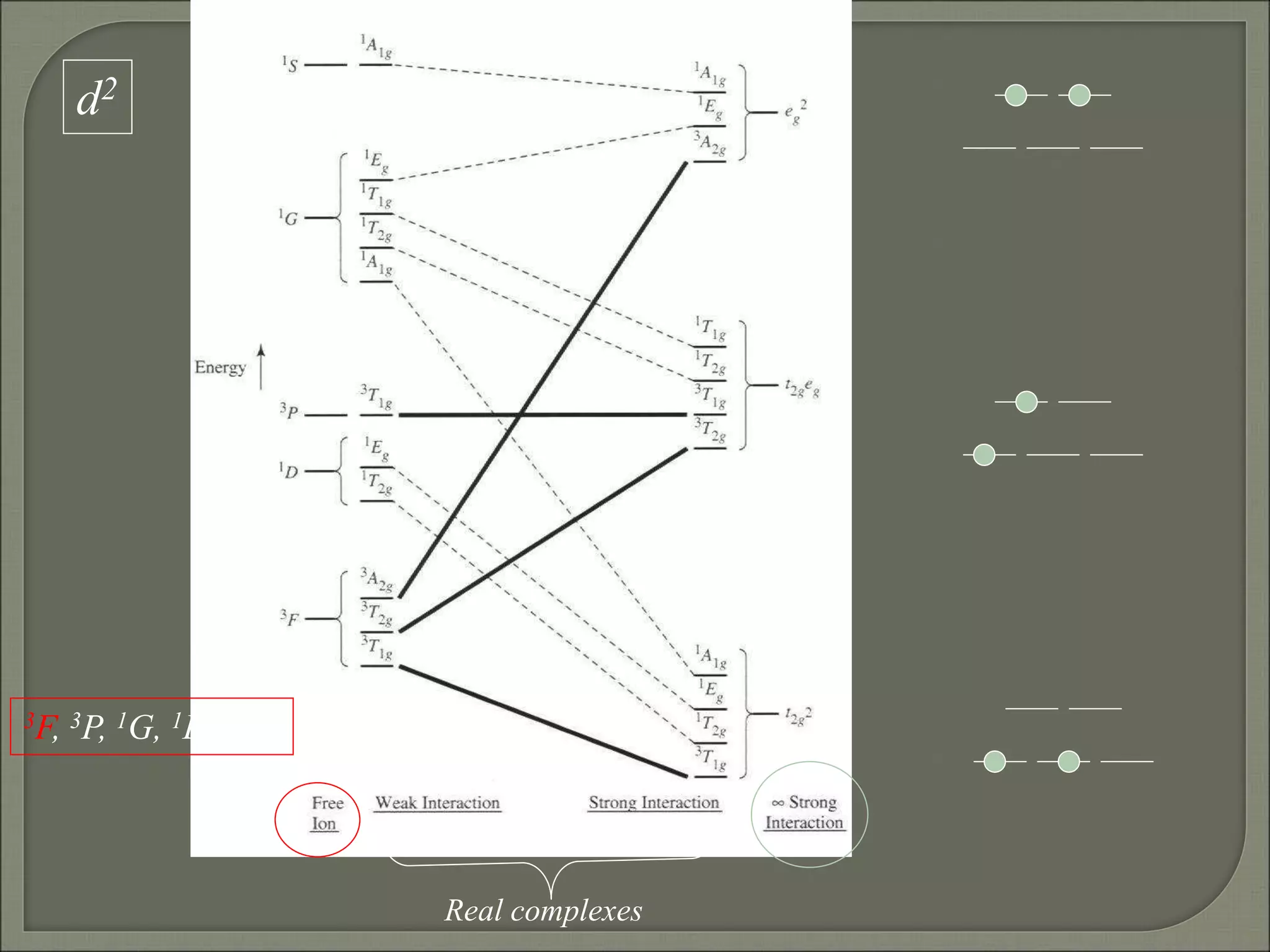
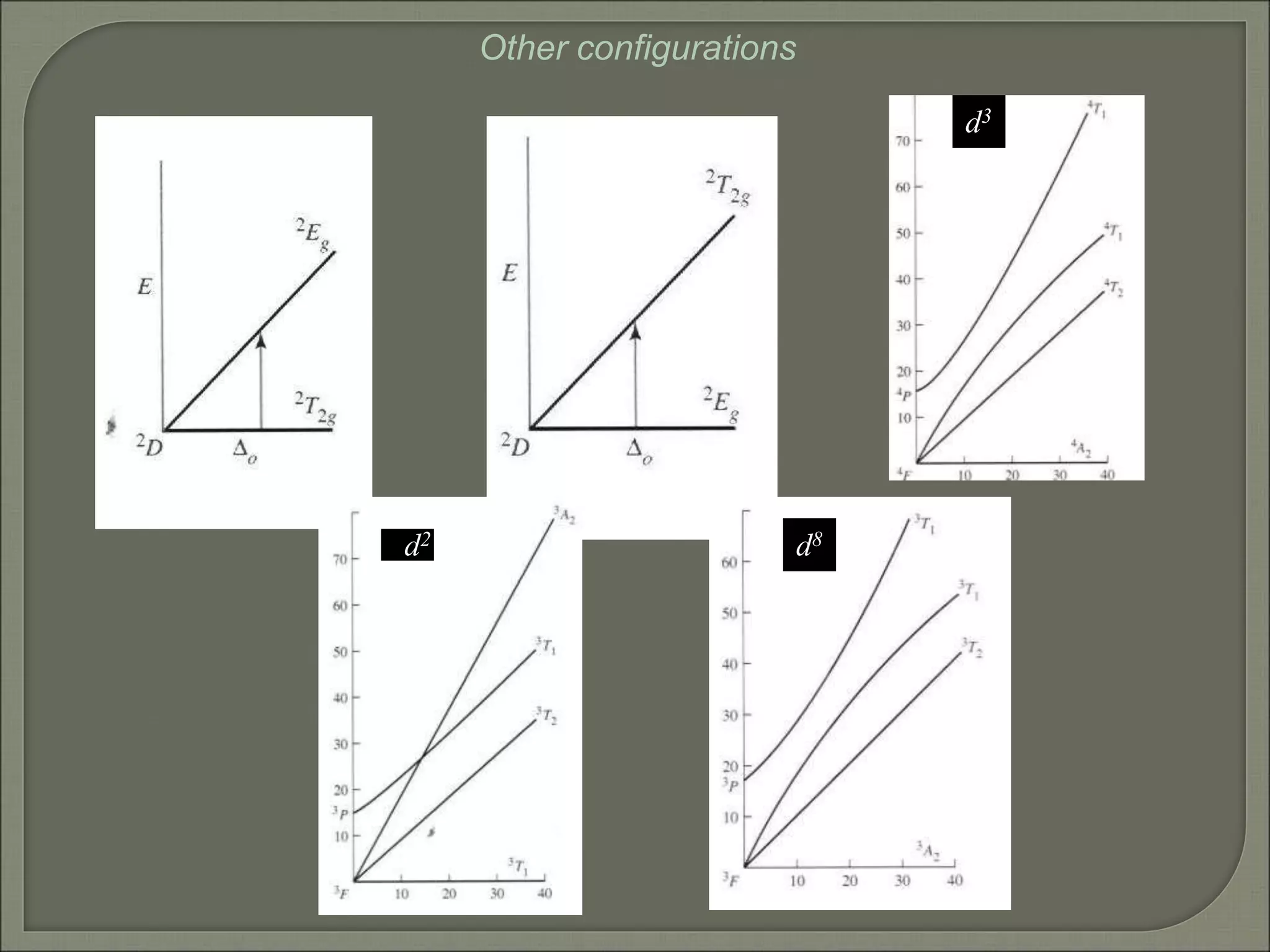
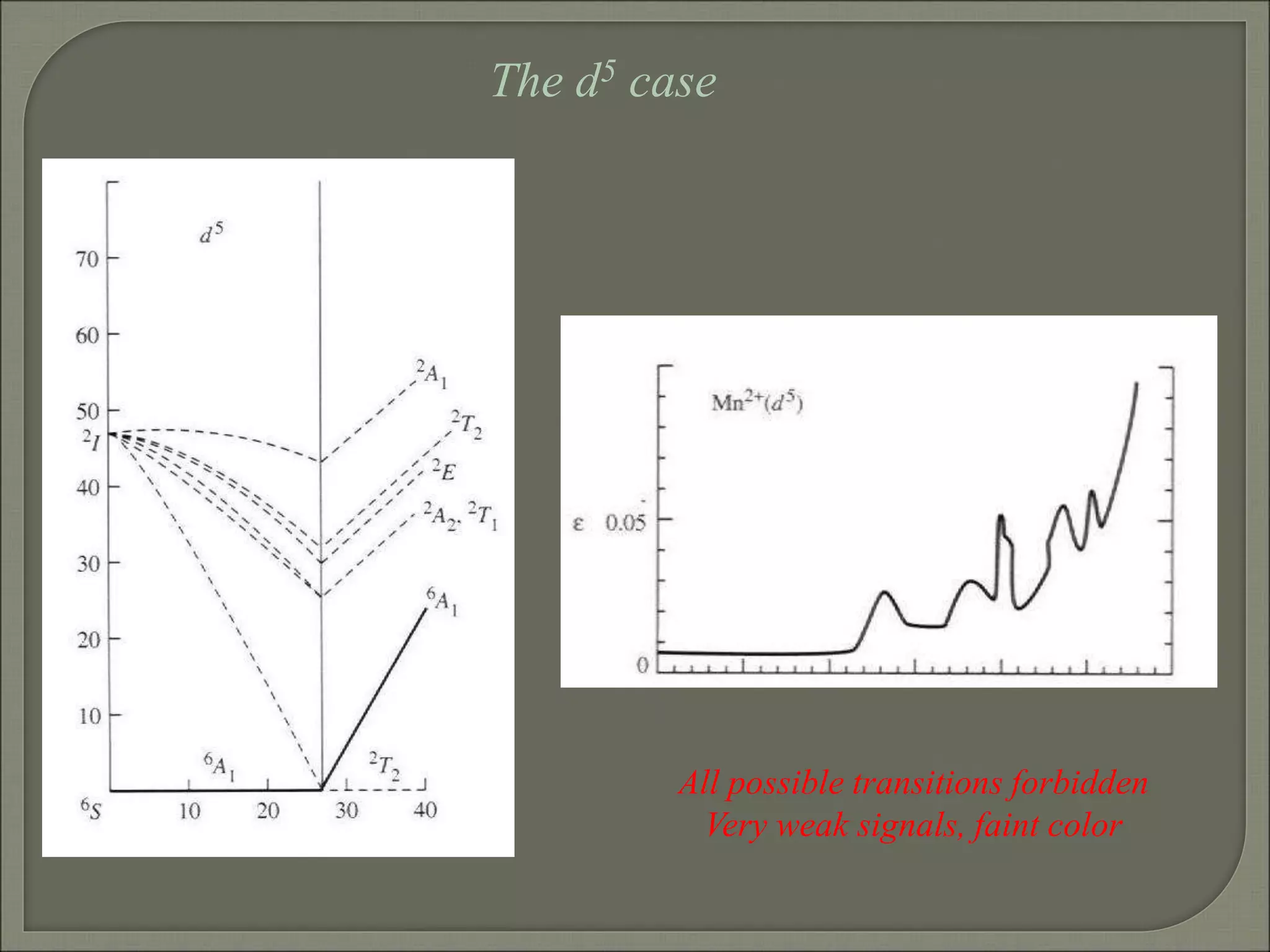
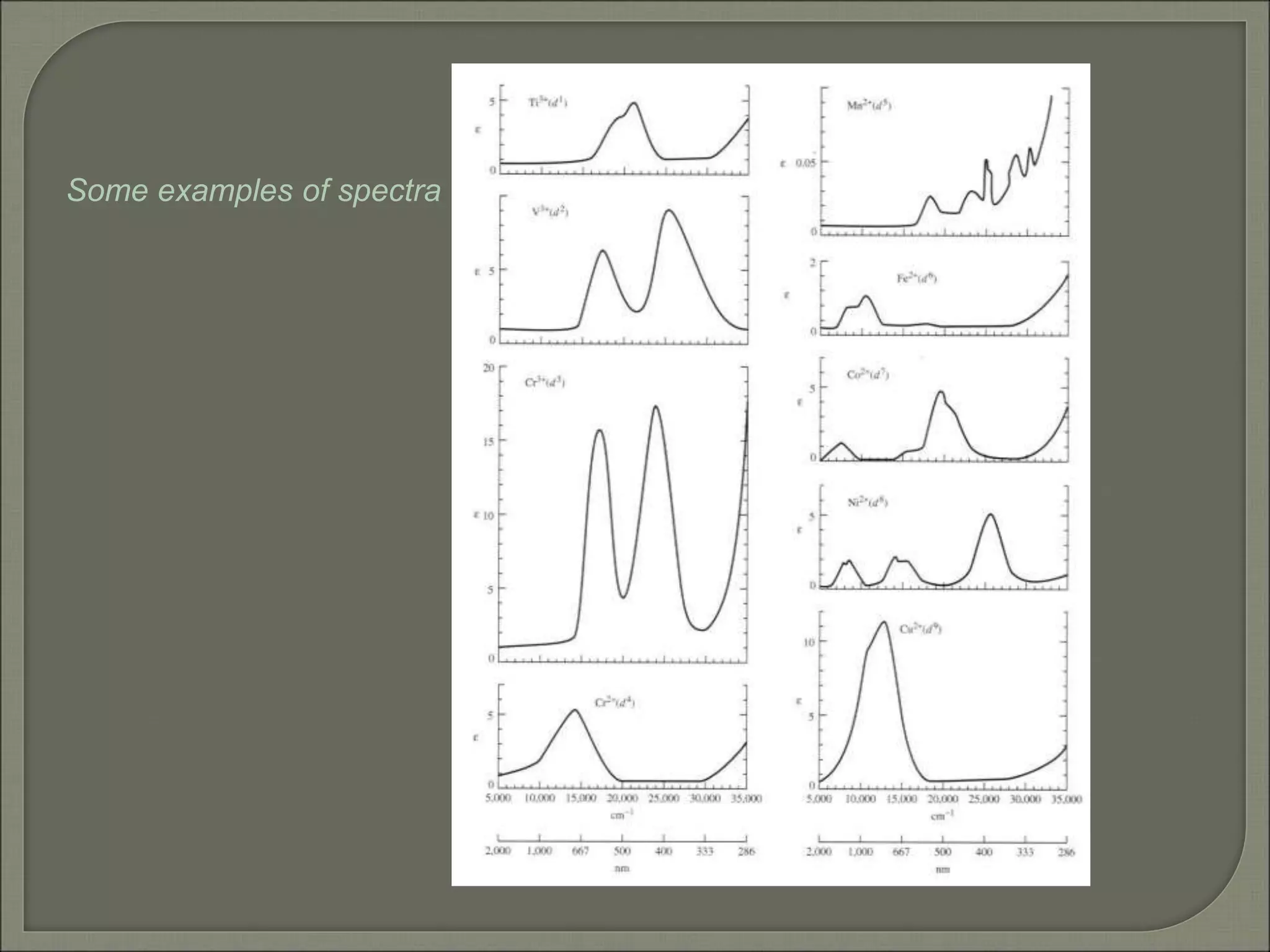
This document discusses the electronic structure and spectra of metal complexes. It begins by introducing ligand field theory and how the d orbitals of the metal ion split into different energy levels depending on the geometry and ligand field strength. Orgel diagrams are used to illustrate the splitting patterns for different d electron configurations from d1 to d10. Selection rules for electronic transitions are described. Tanabe-Sugano diagrams show how transition energies vary with ligand field strength. Methods for determining the ligand field splitting parameter (Δo) from experimental spectra are also outlined, along with examples of different types of spectra observed.