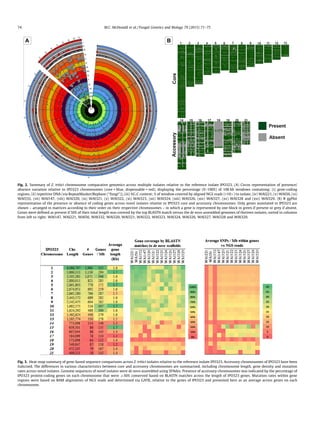
This document describes a study that uses next-generation re-sequencing and bioinformatics to analyze presence/absence variation of accessory chromosomes across isolates of the wheat pathogen Zymoseptoria tritici. The genome of the reference isolate IPO323 contains 21 chromosomes including 8 accessory chromosomes. Low-cost next-generation sequencing of 13 novel Z. tritici isolates is performed and the reads are aligned to the IPO323 reference genome to determine if accessory chromosomes present in IPO323 are also present in the novel isolates based on read coverage. De novo assembly of reads from the novel isolates is also conducted and compared to IPO323 to identify any additional accessory chromosomes or sequences not present in IPO323. This