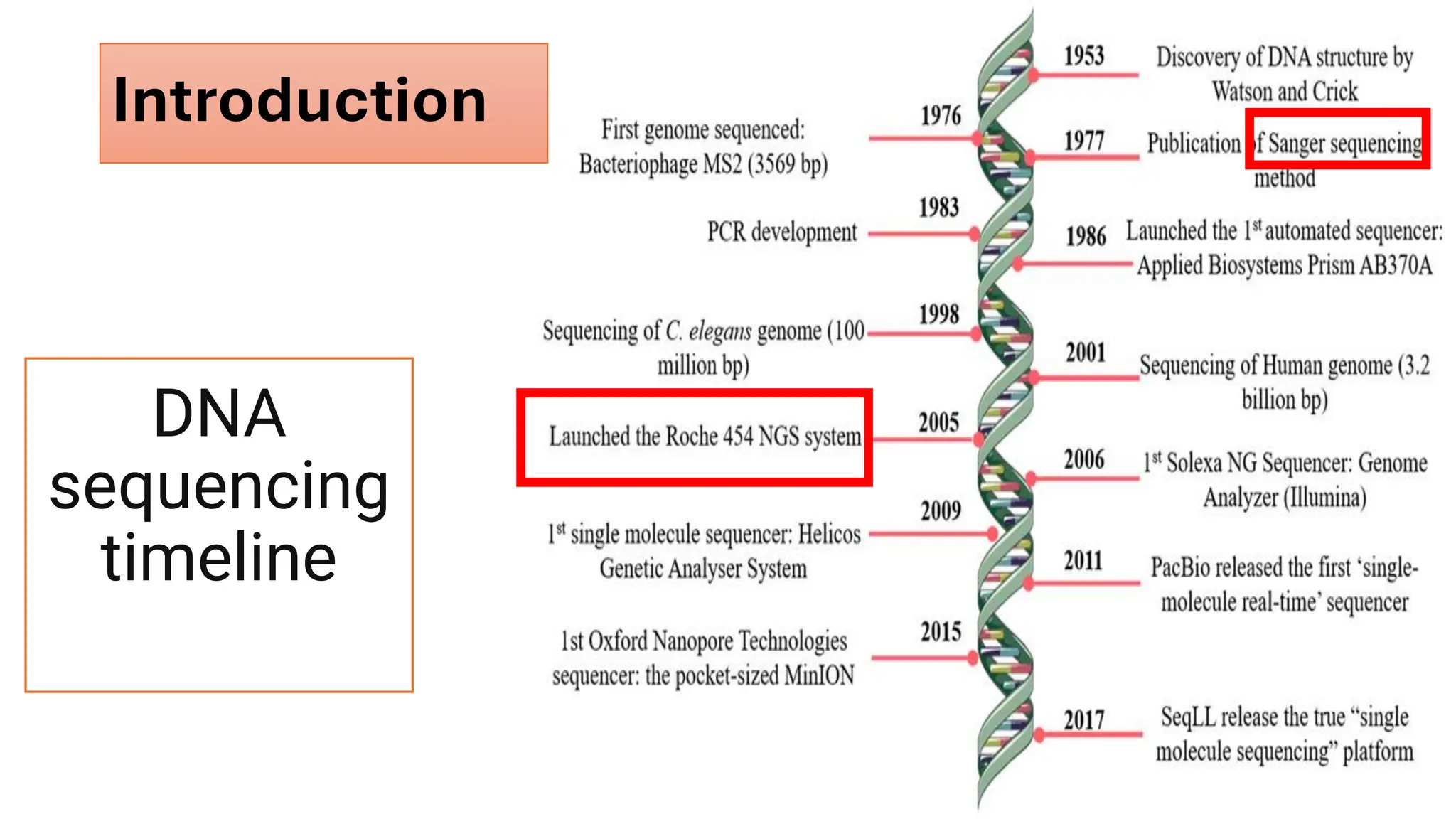
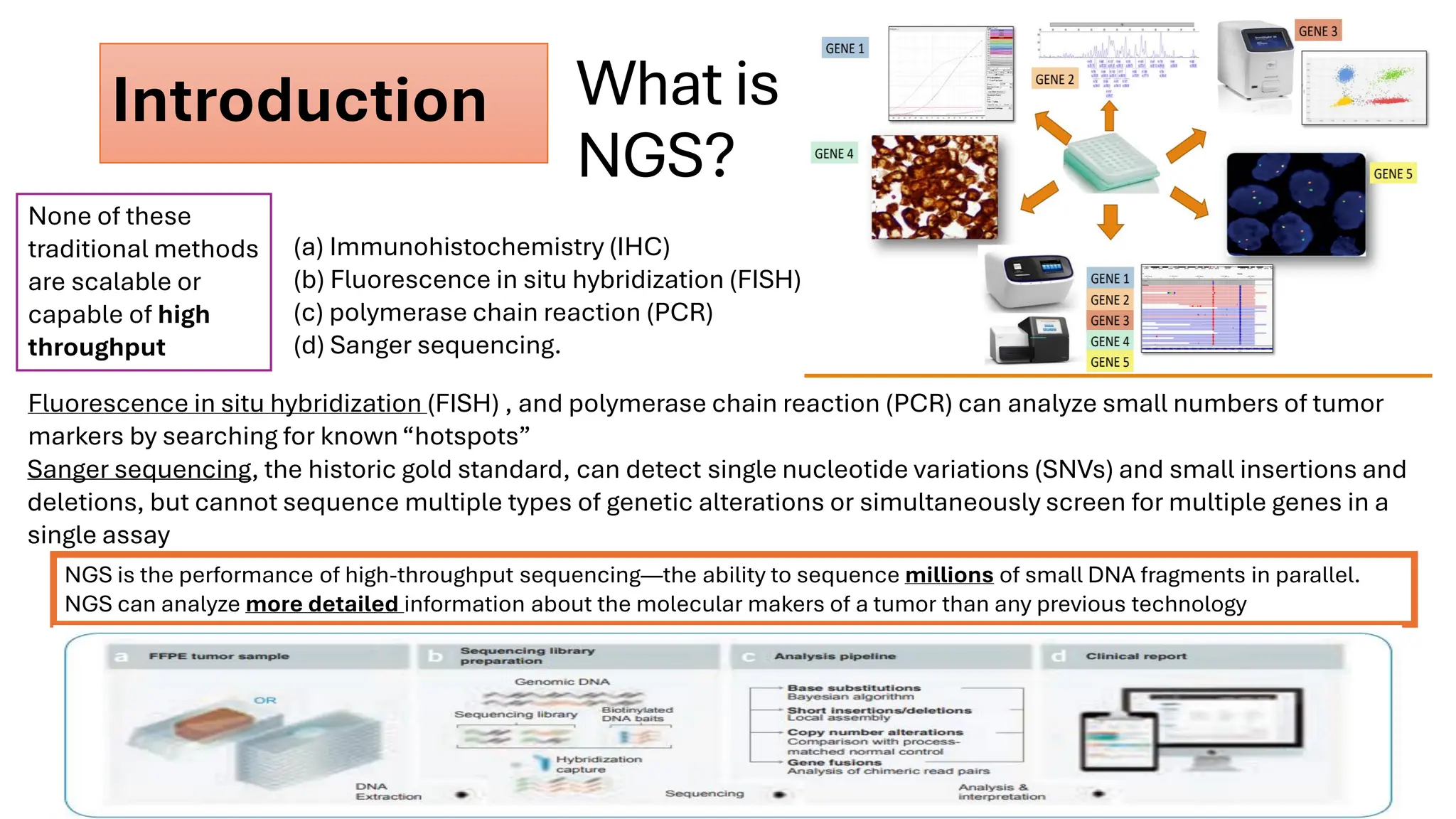
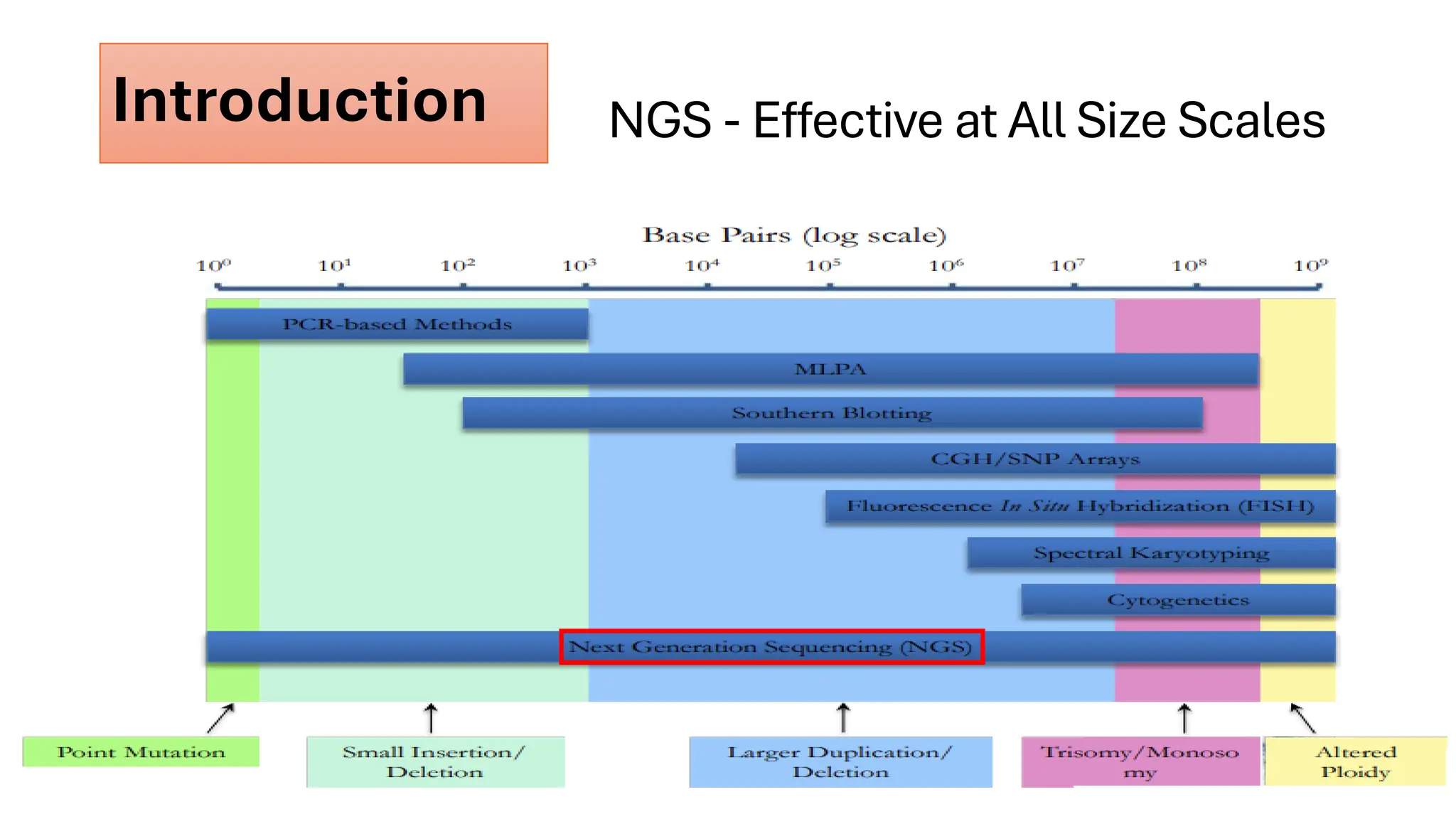
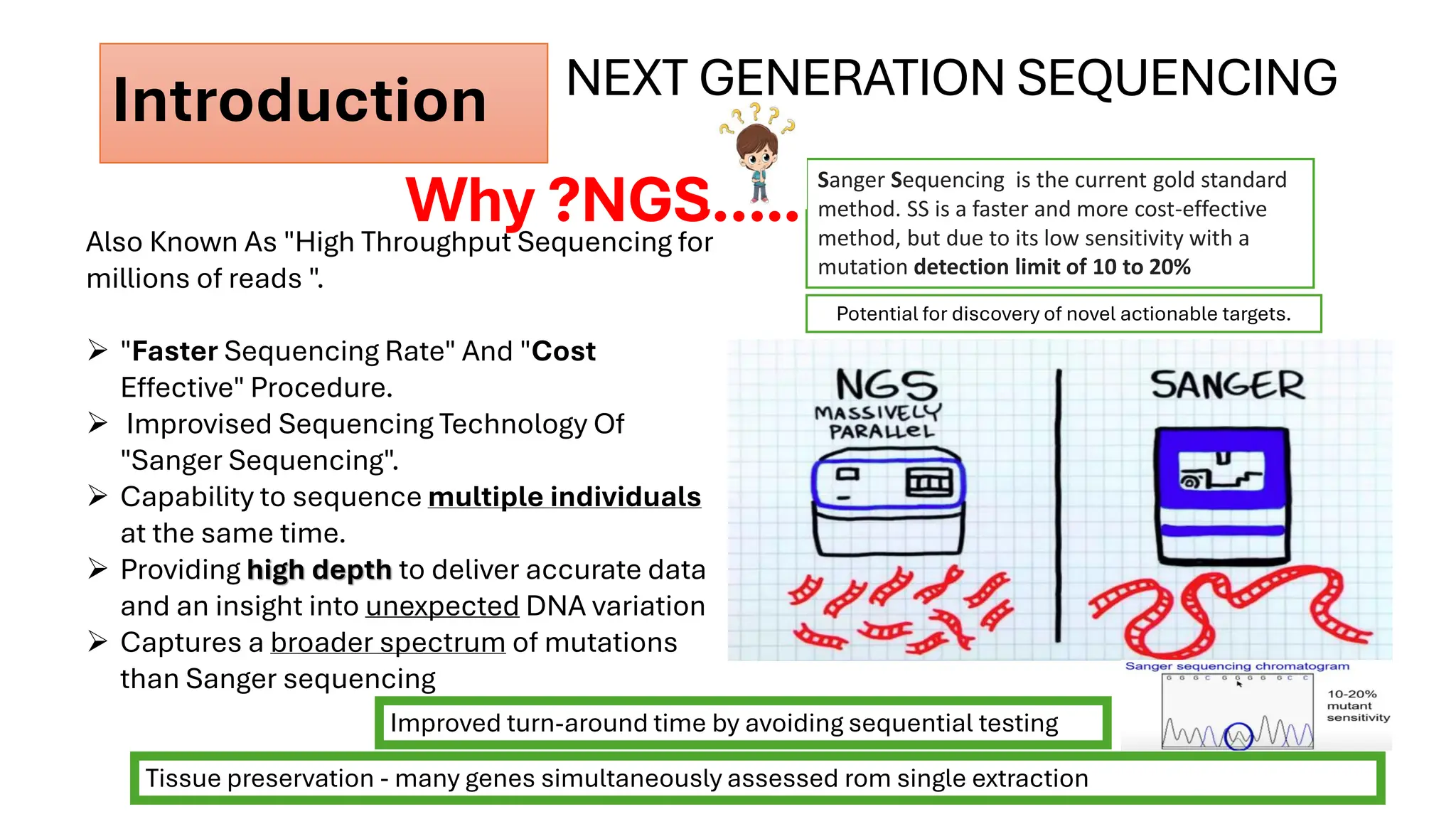
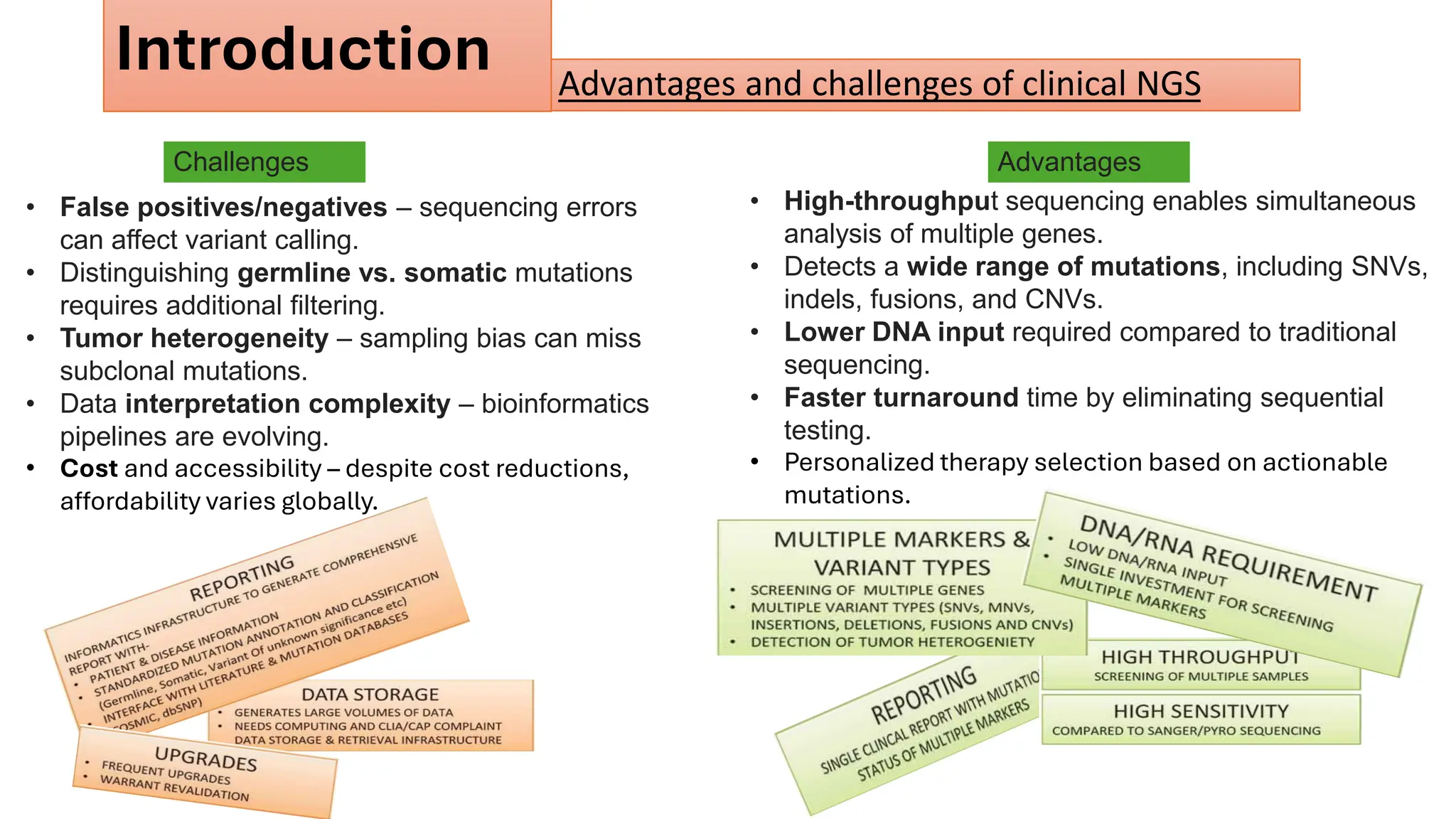
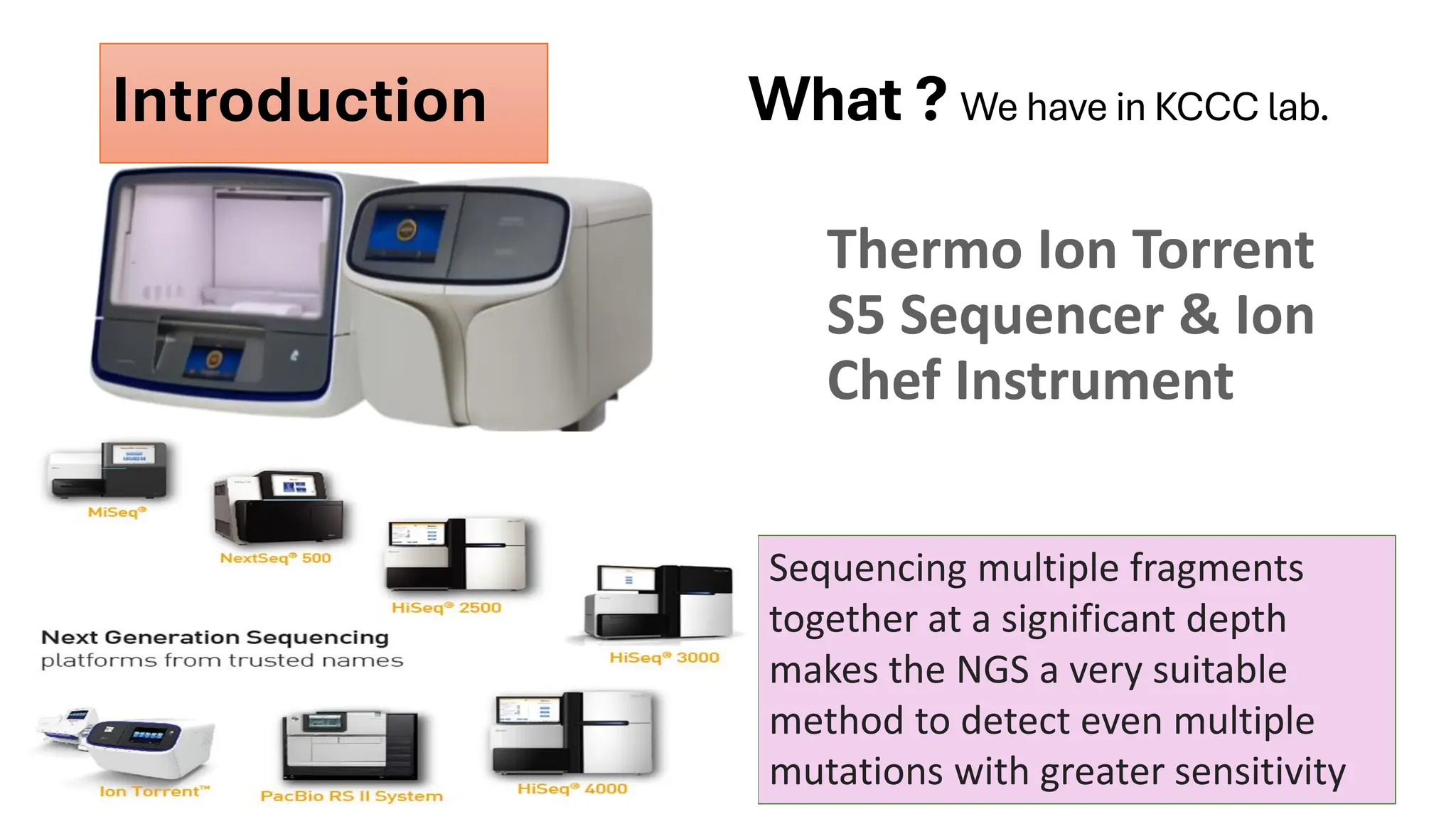
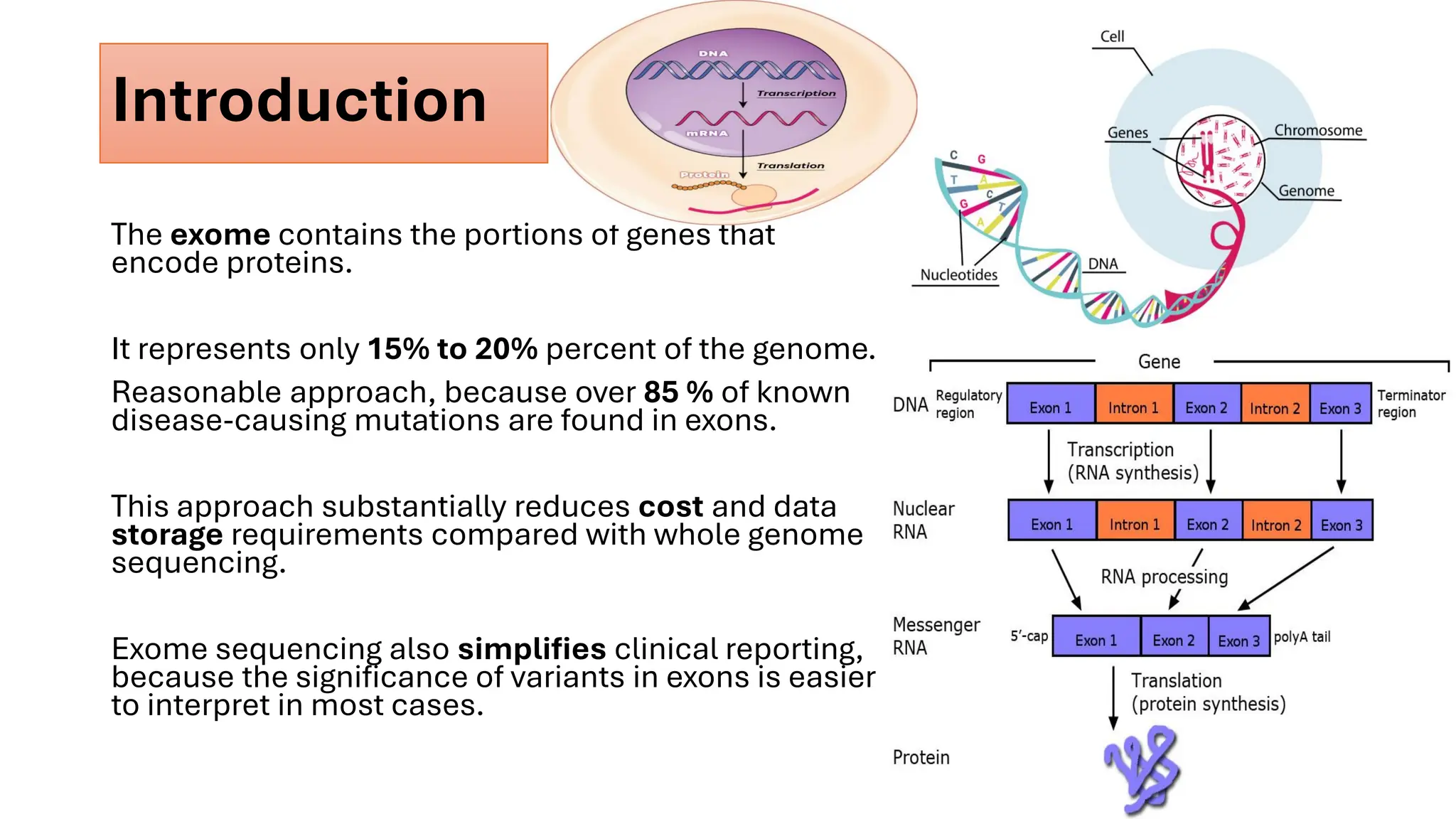
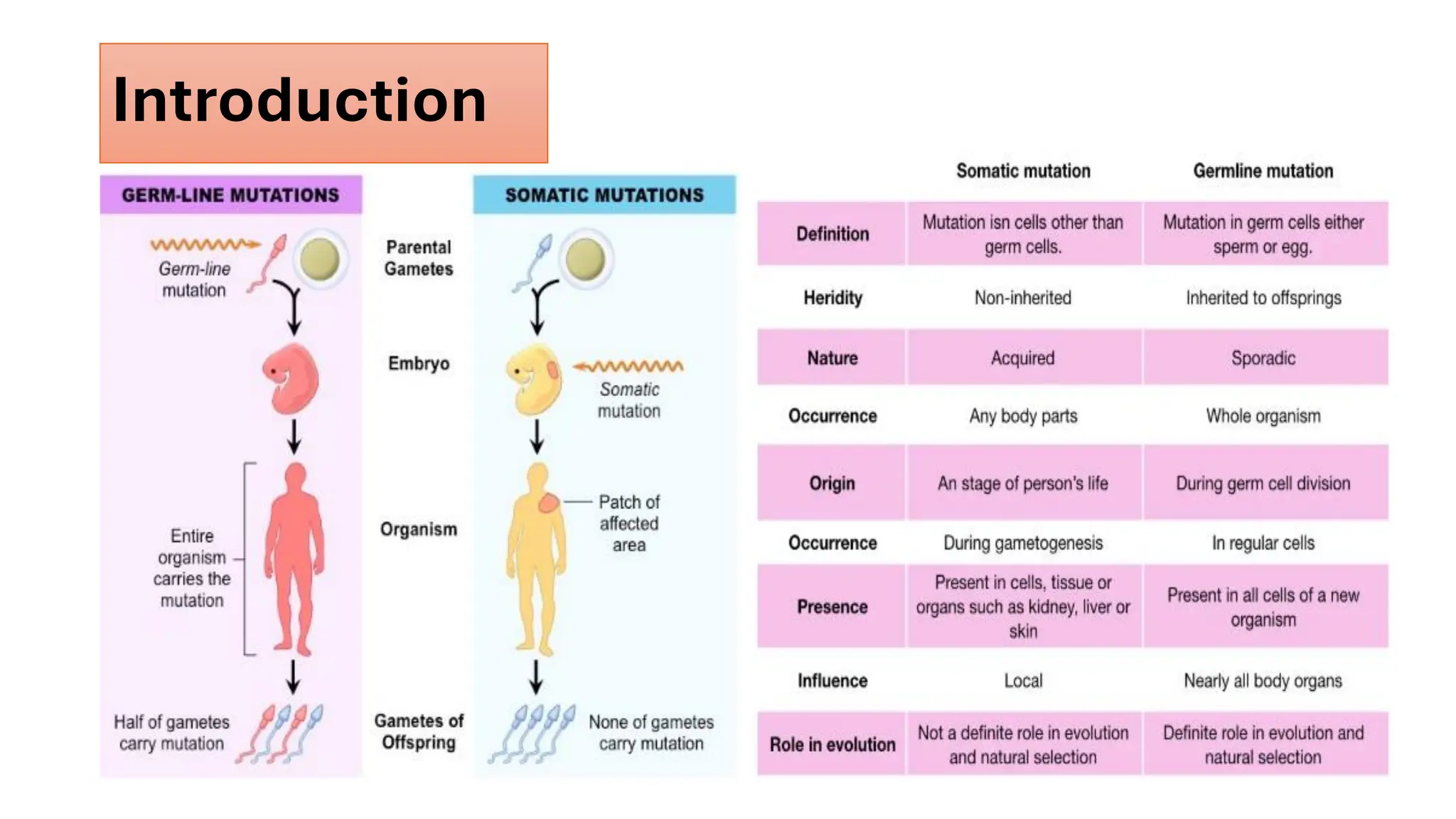
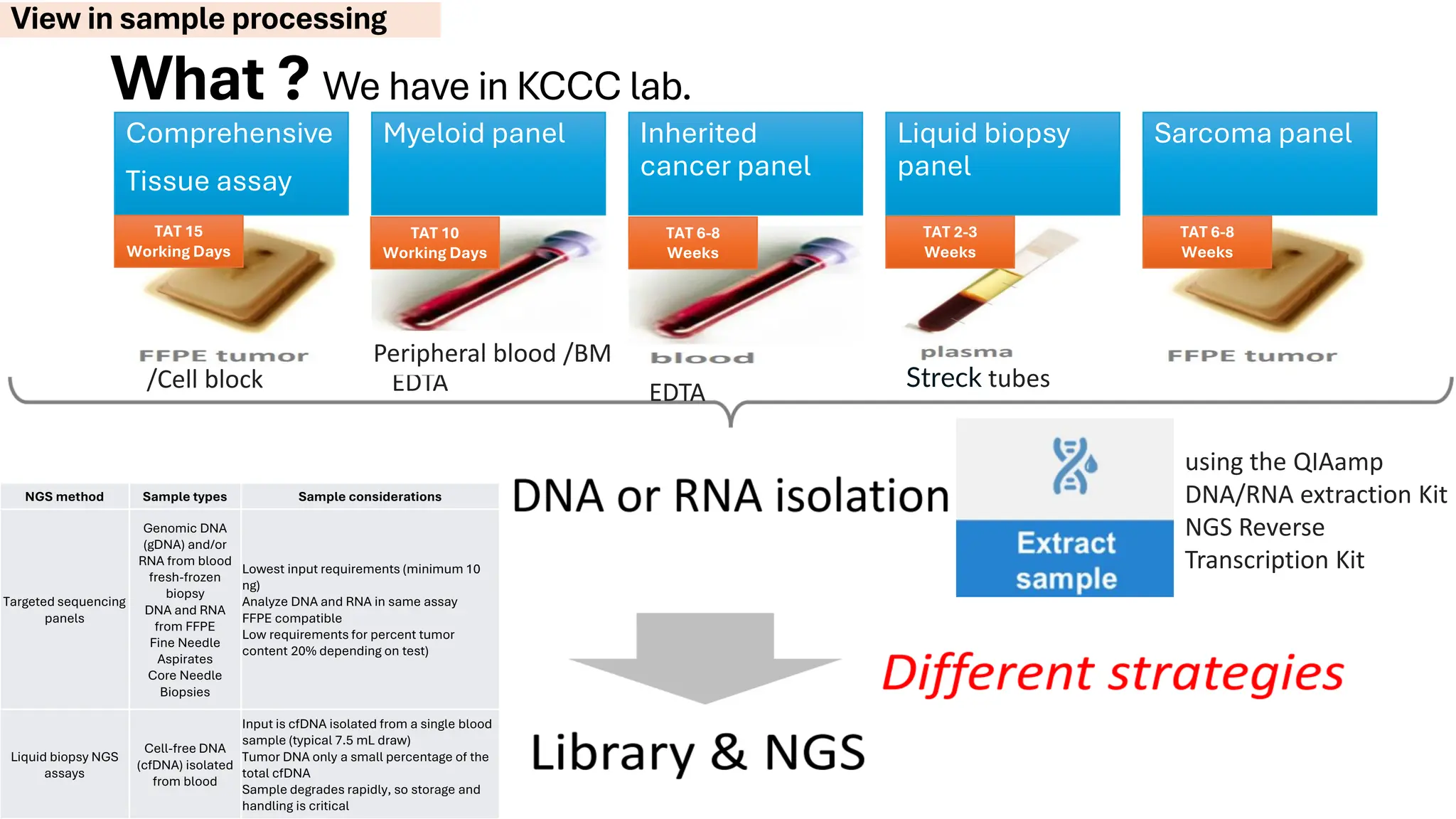
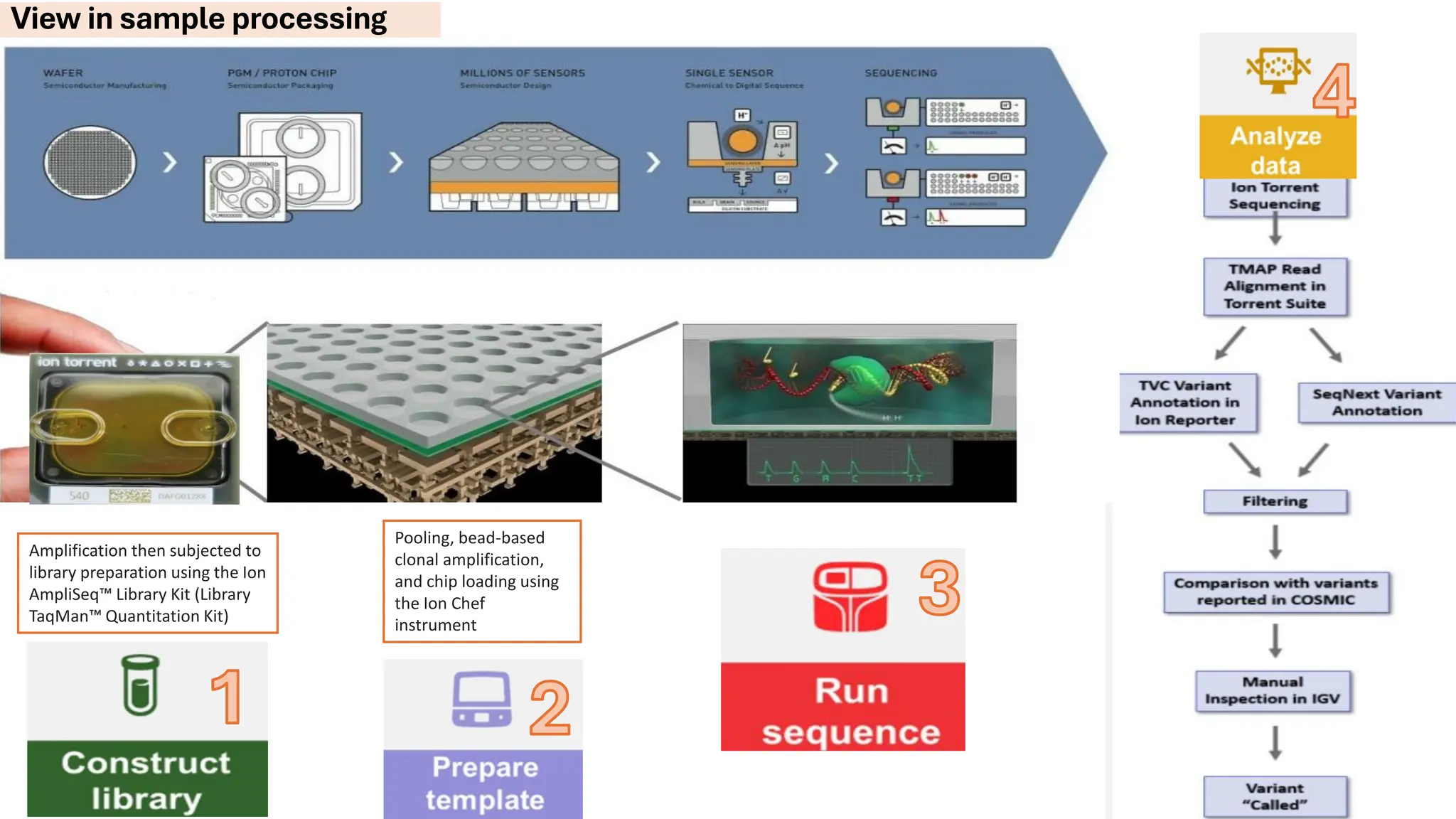
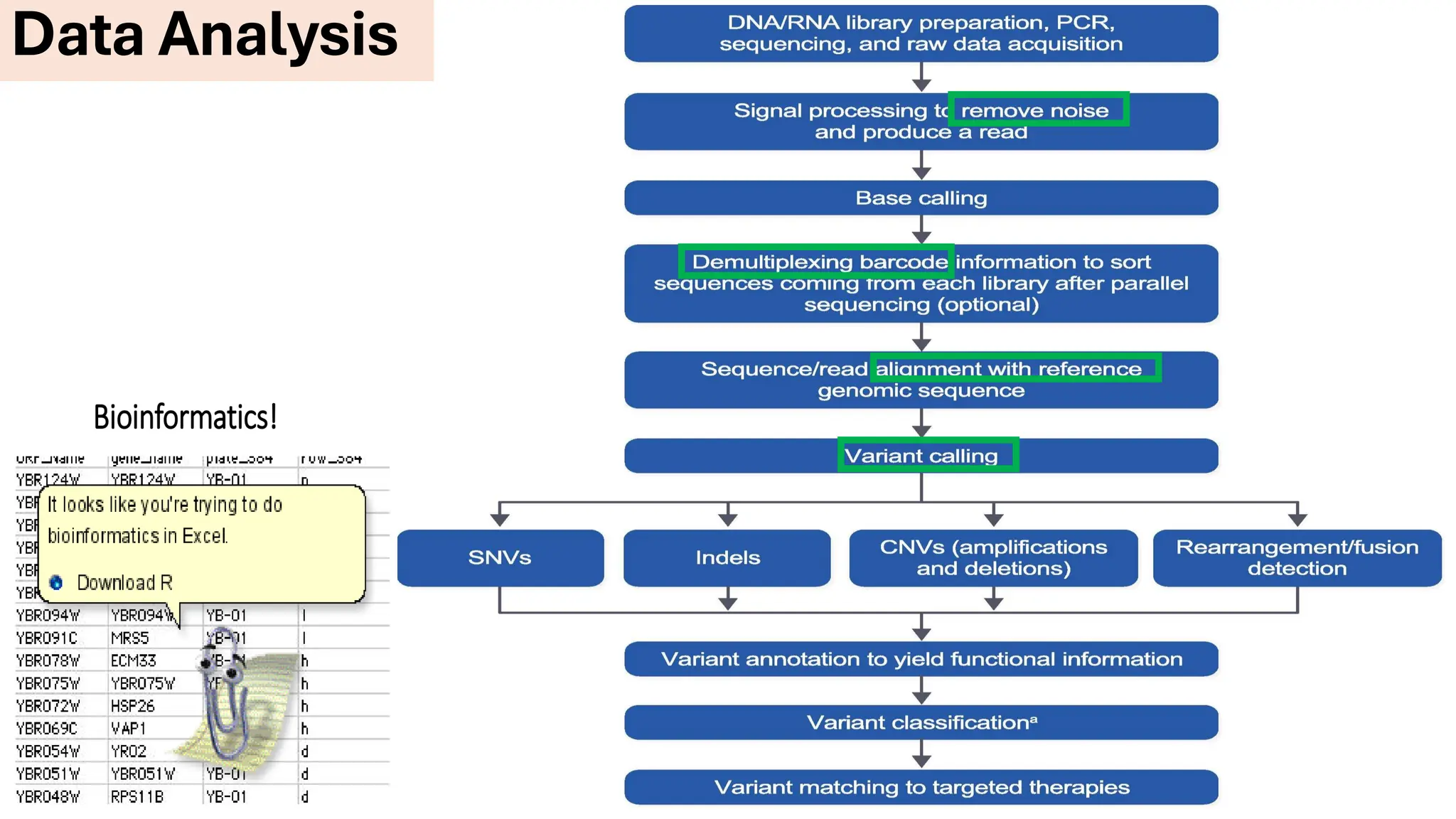
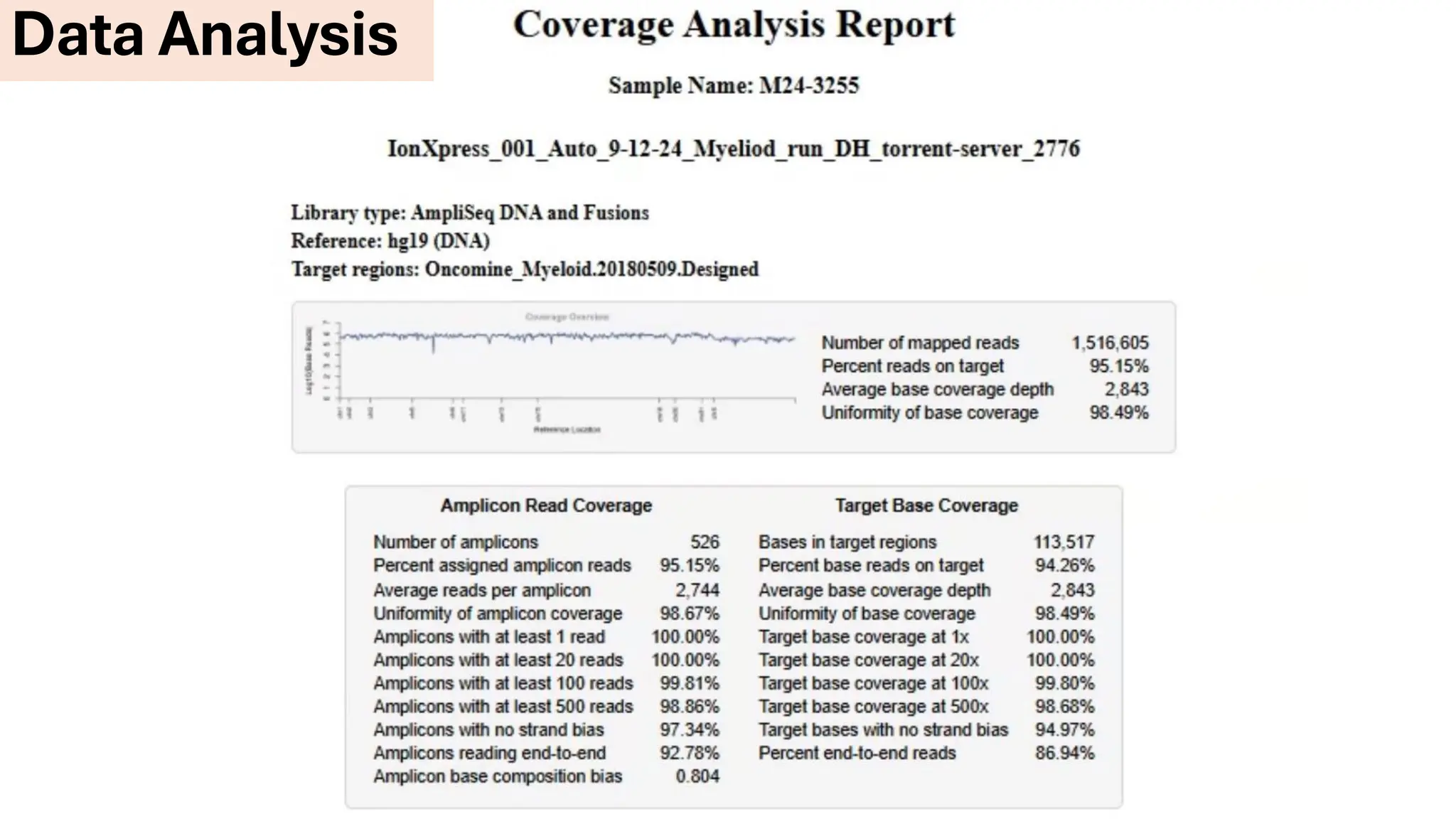
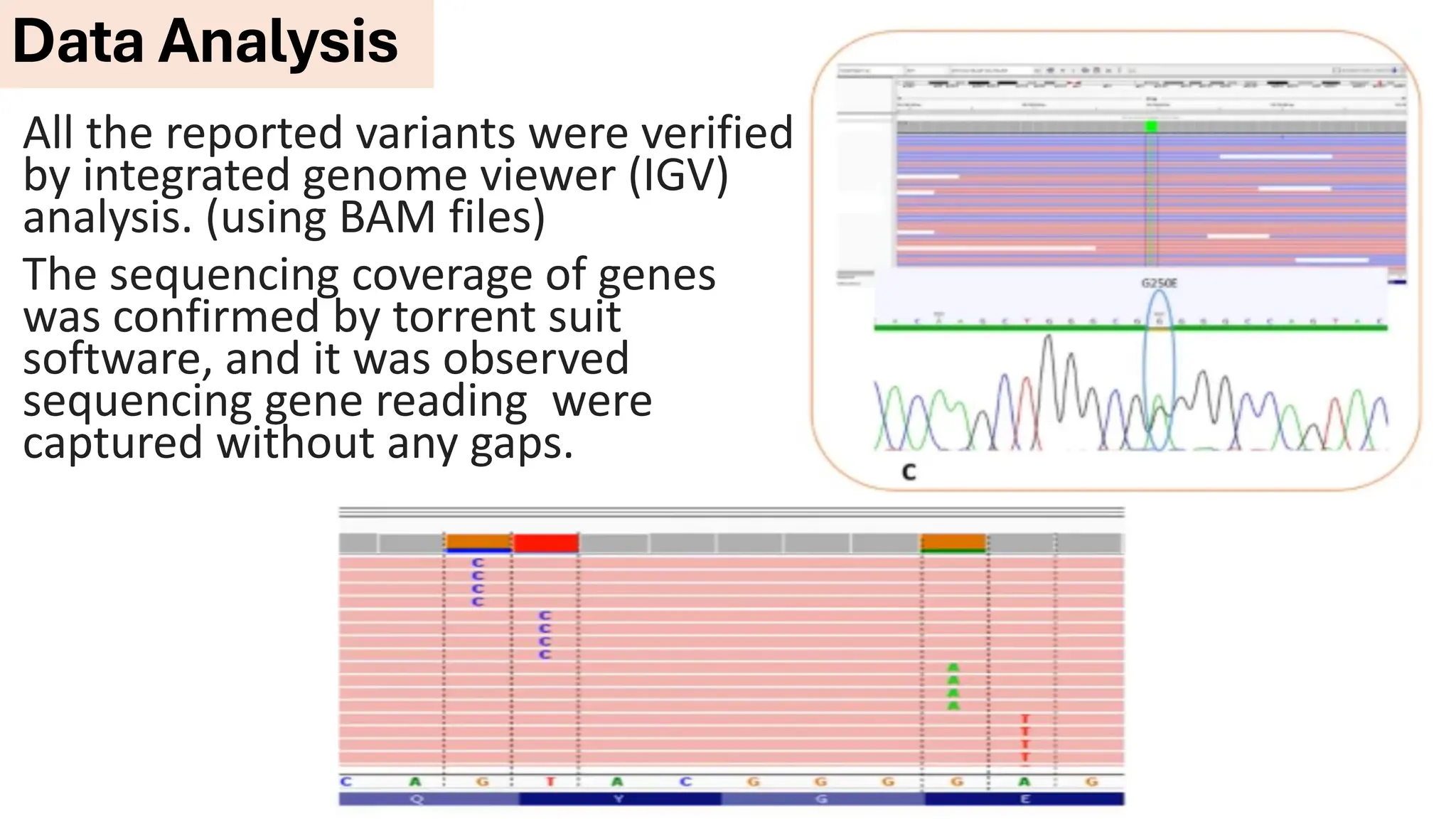
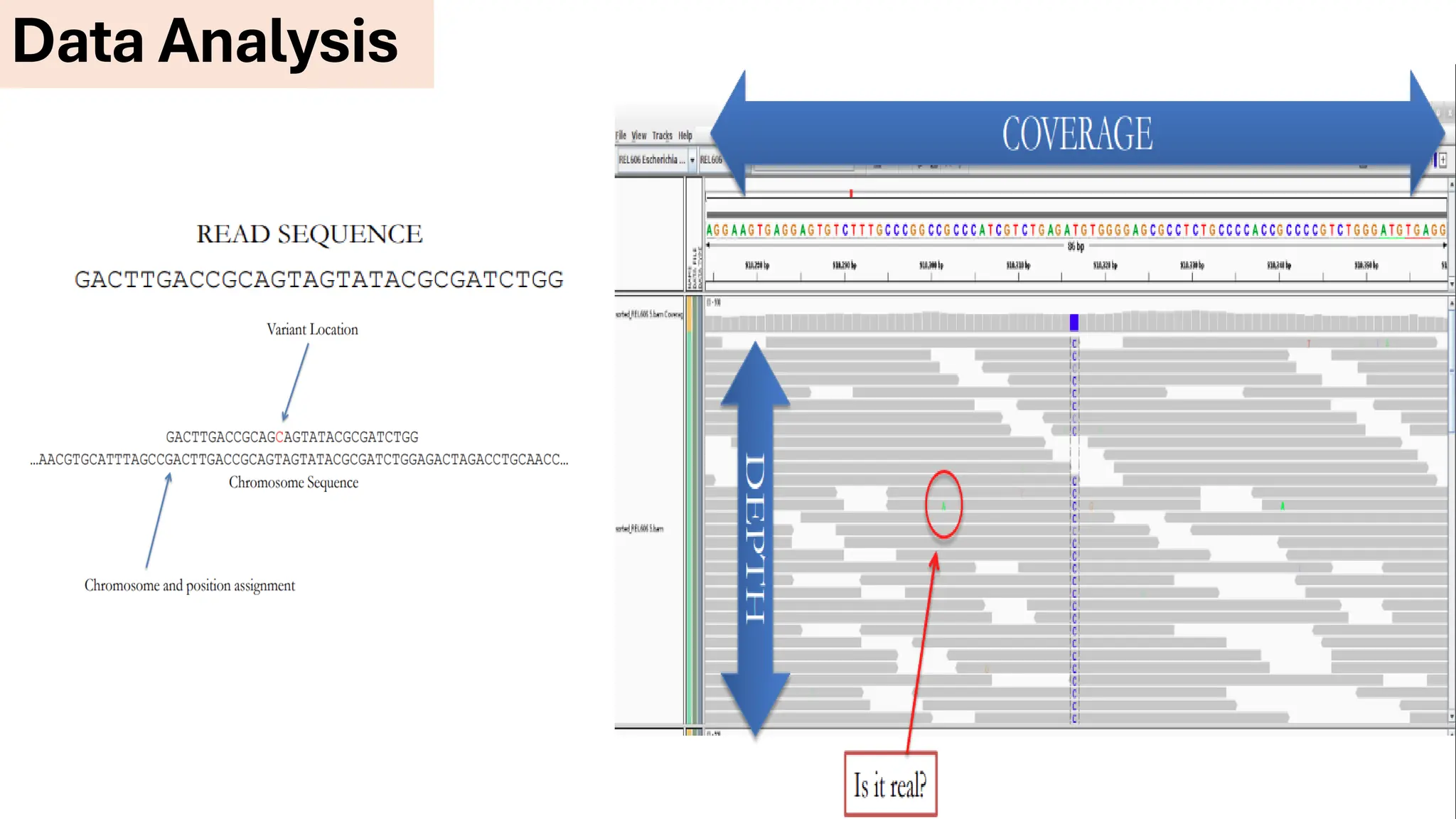
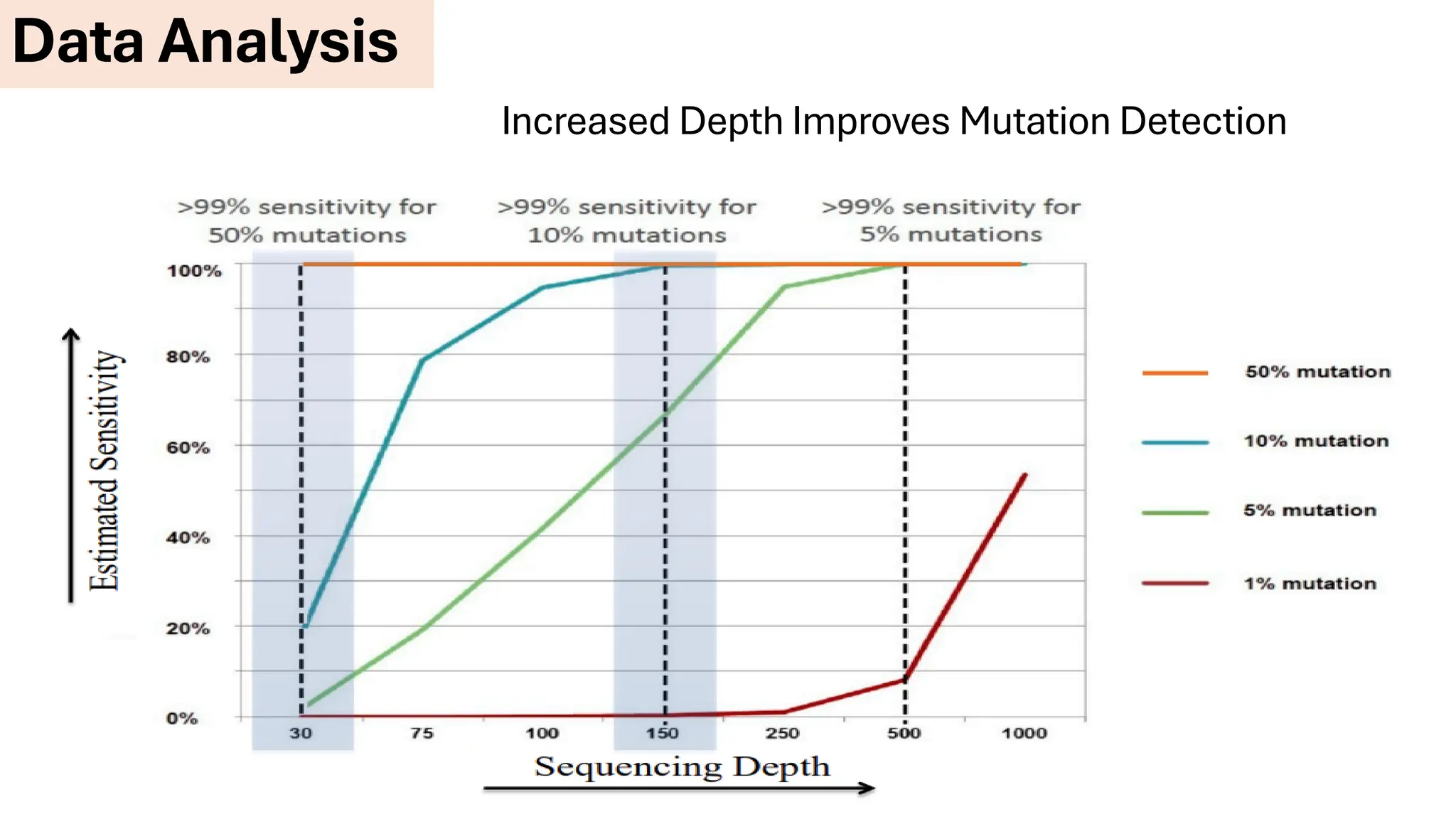
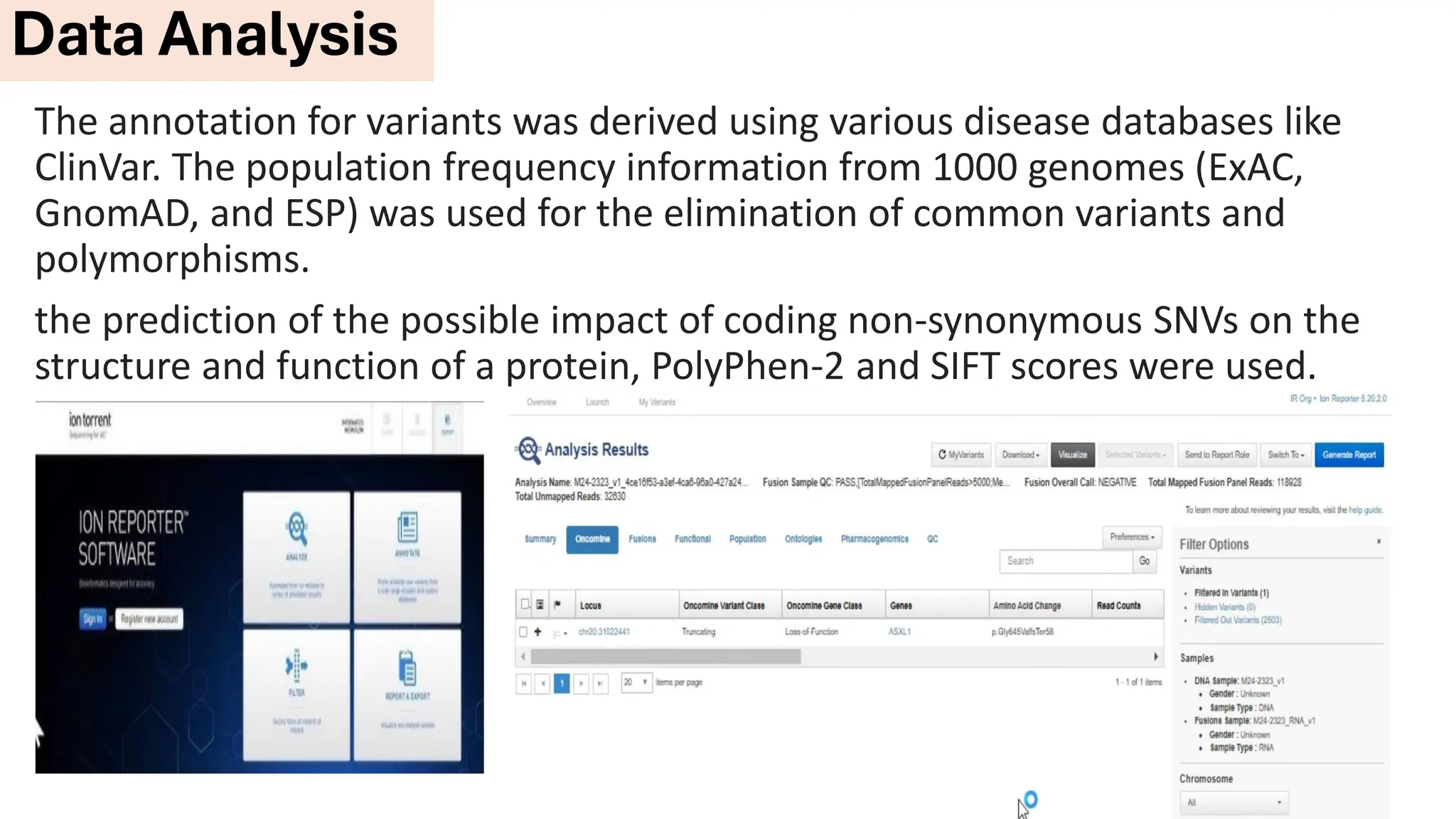
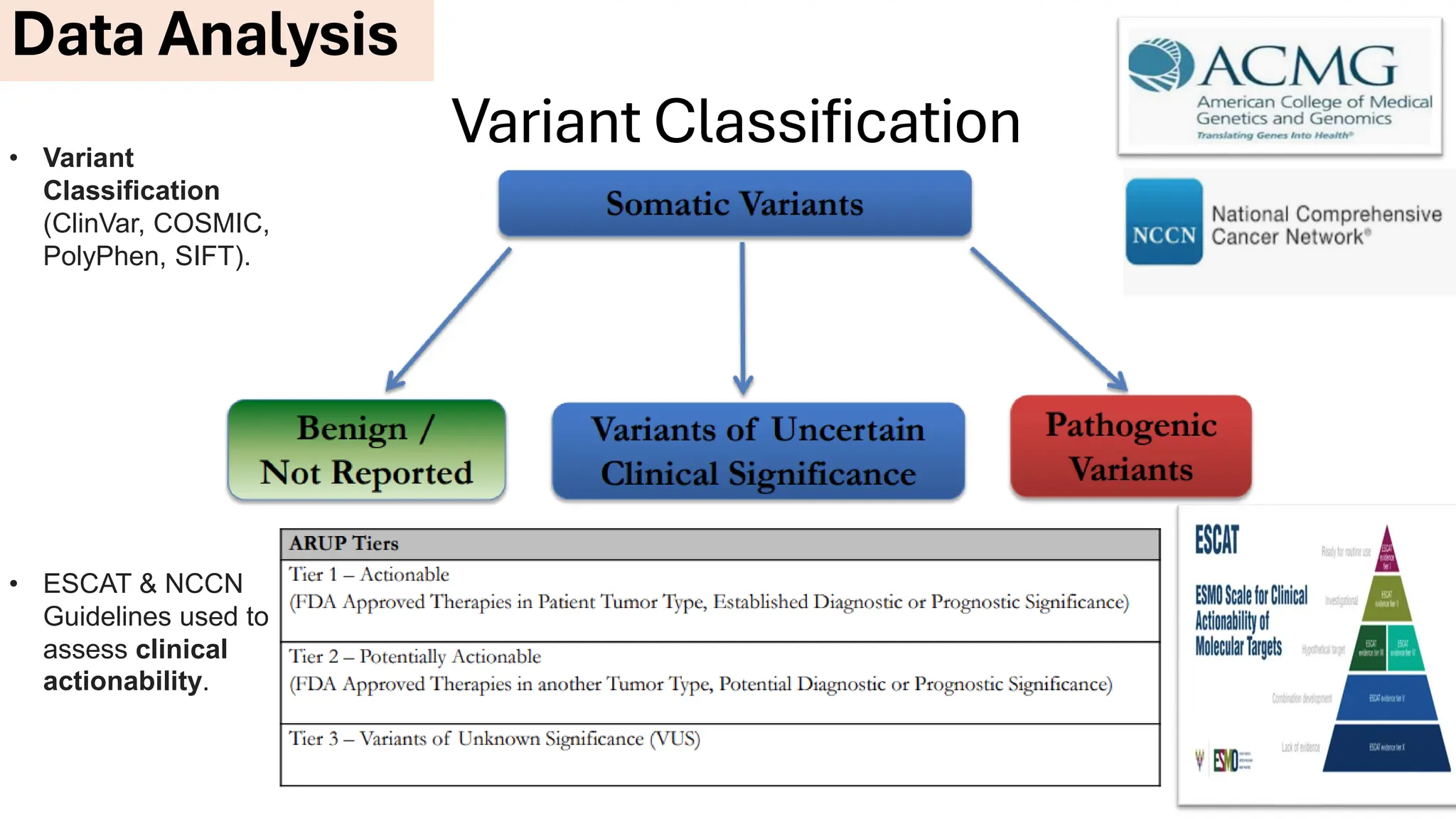
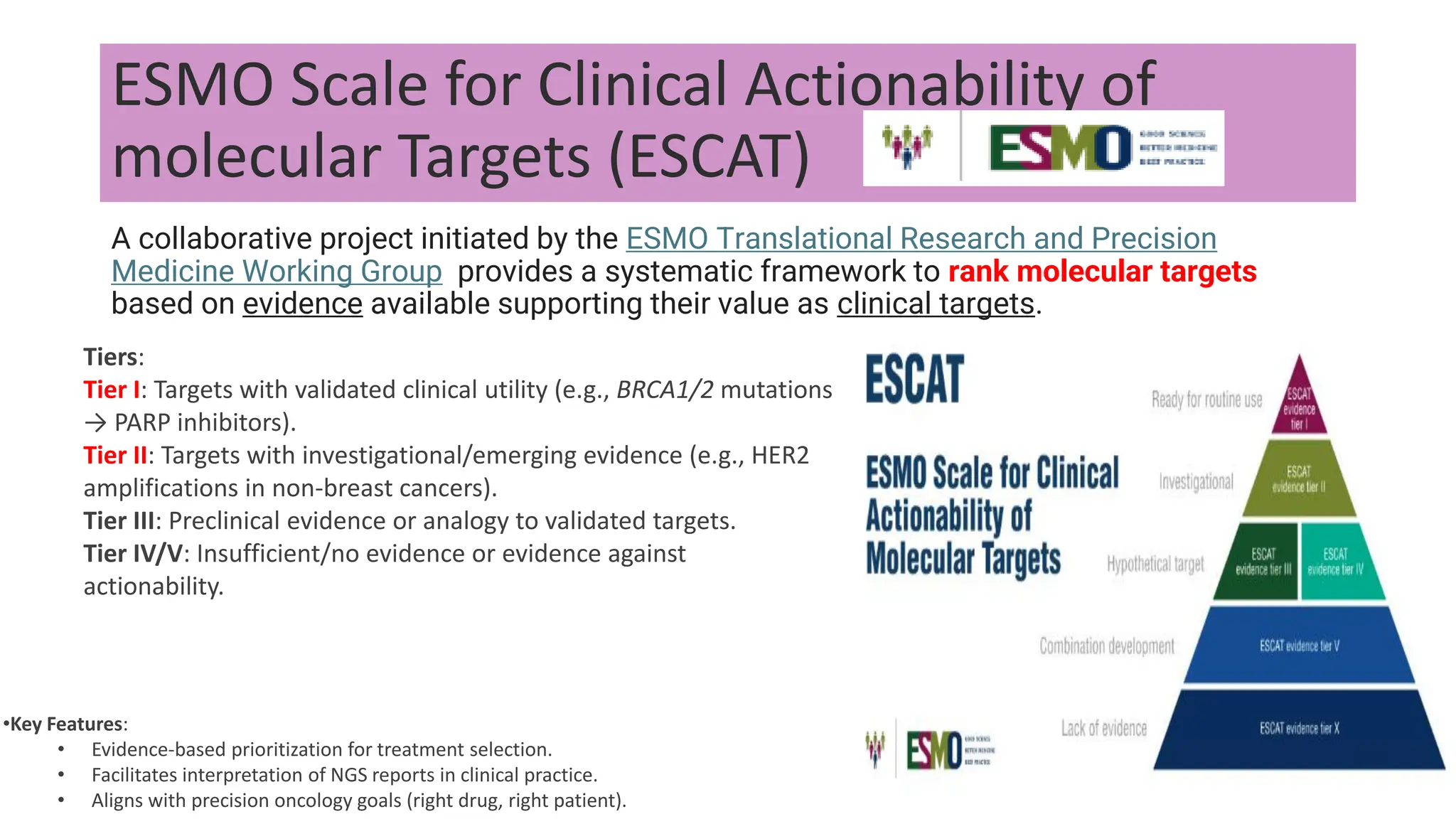
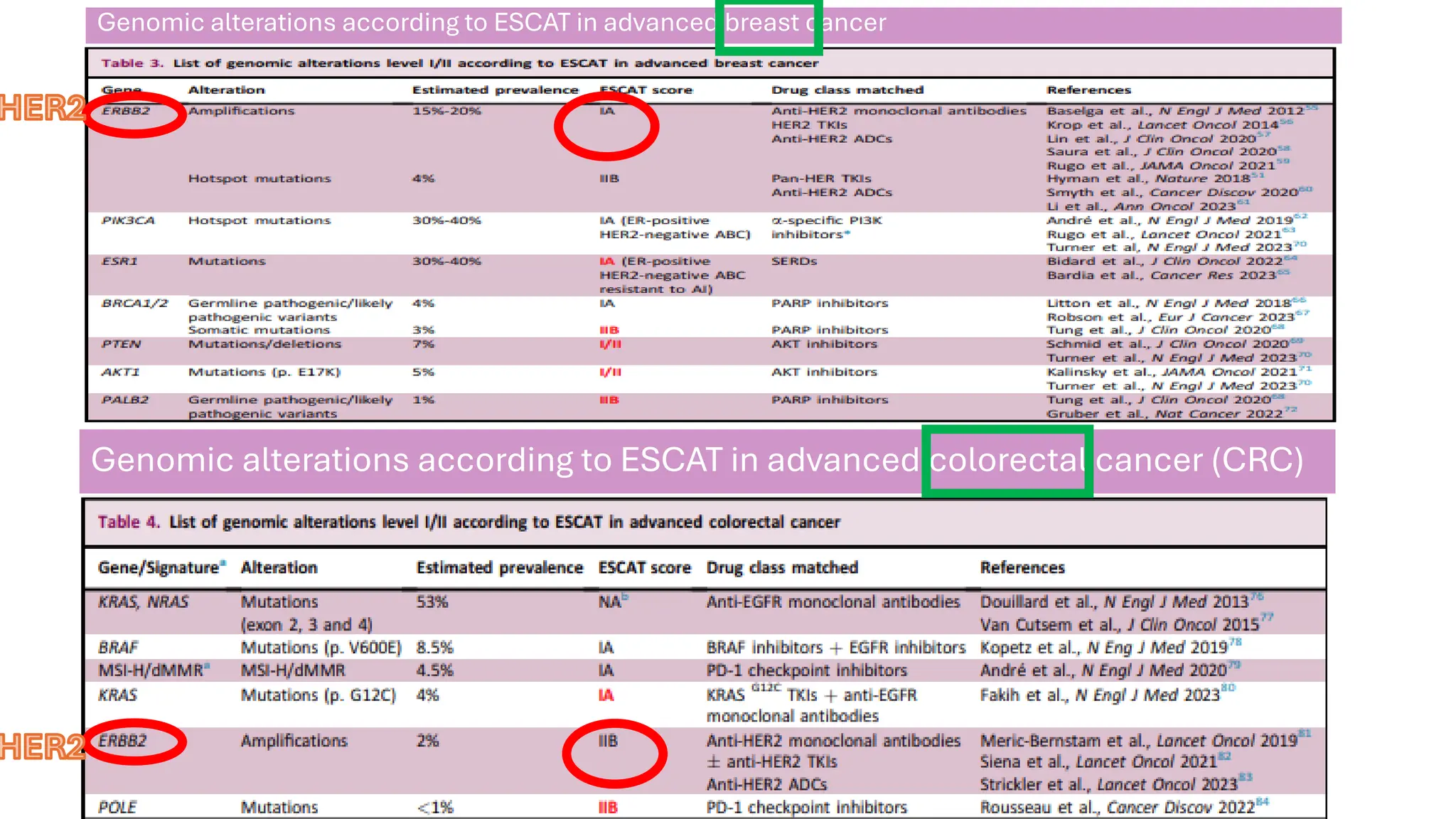
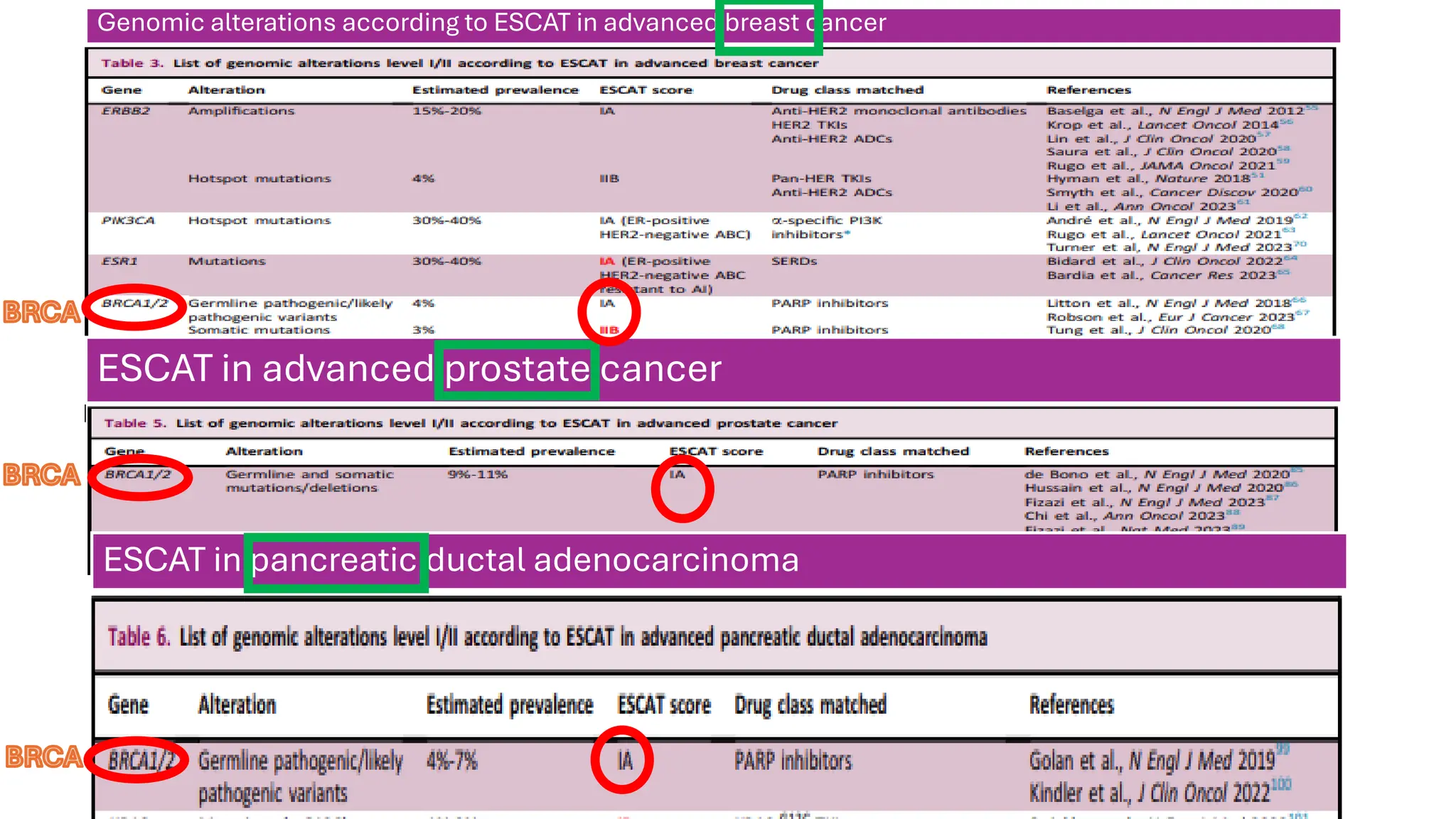
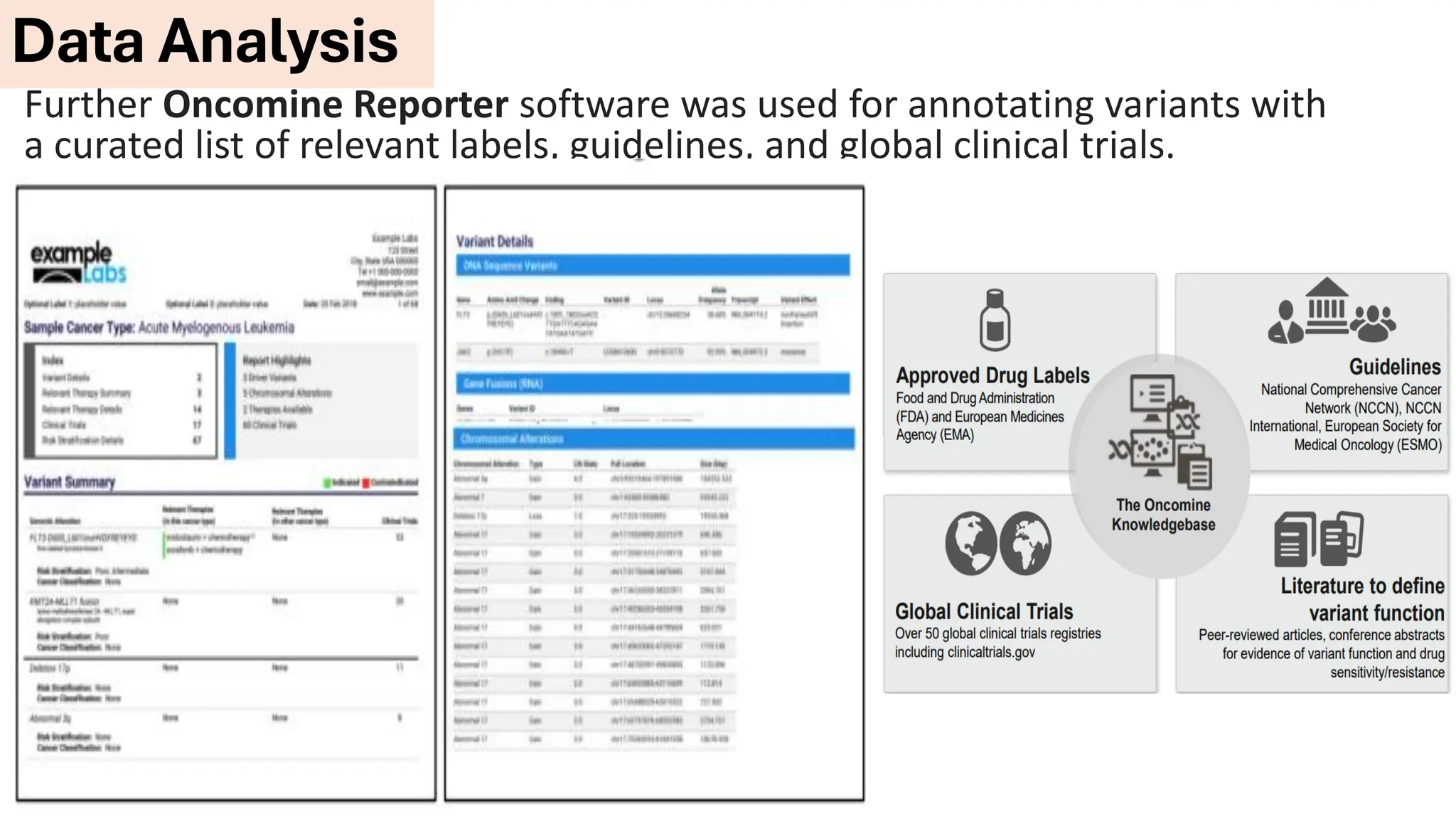
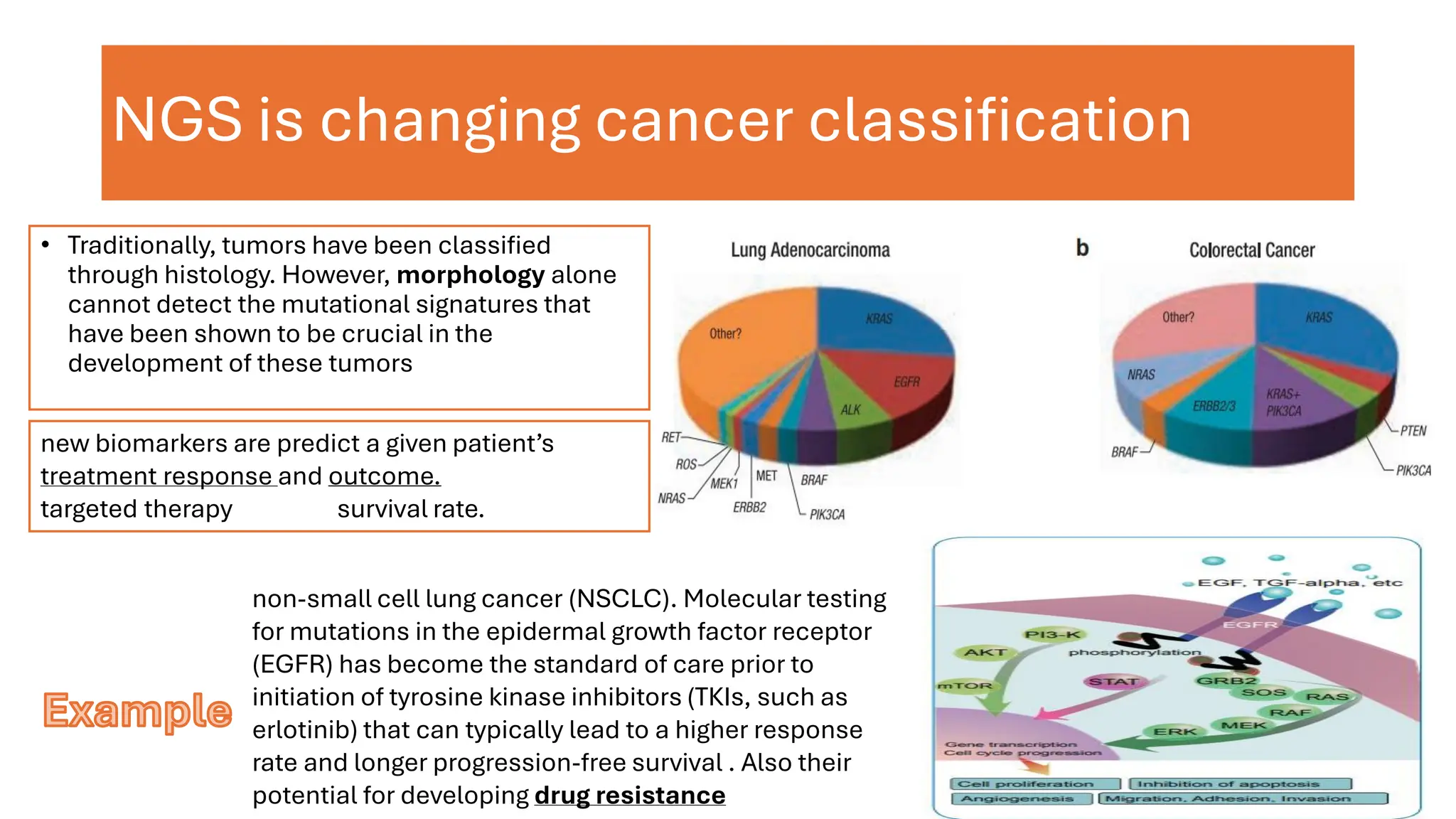
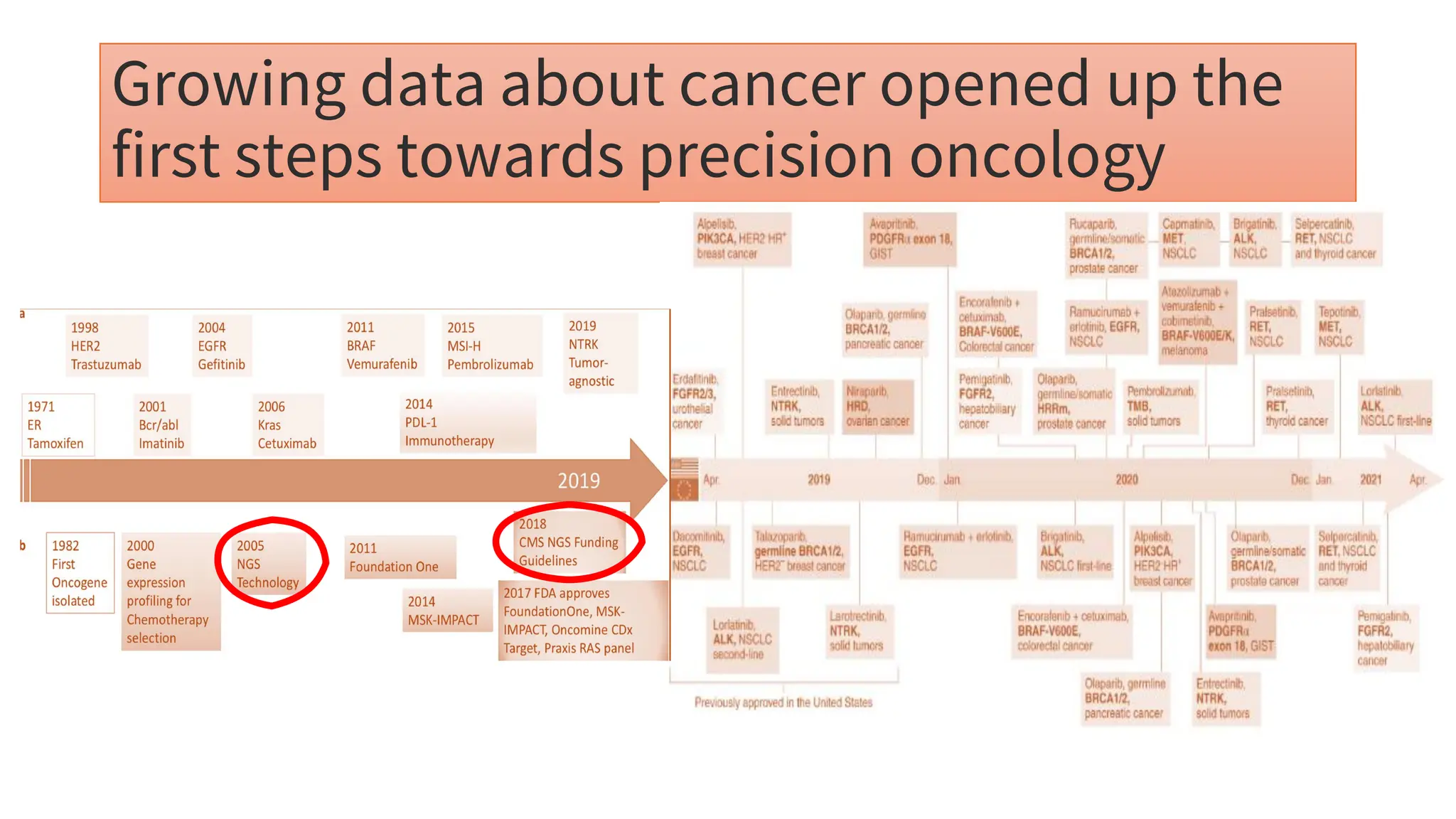
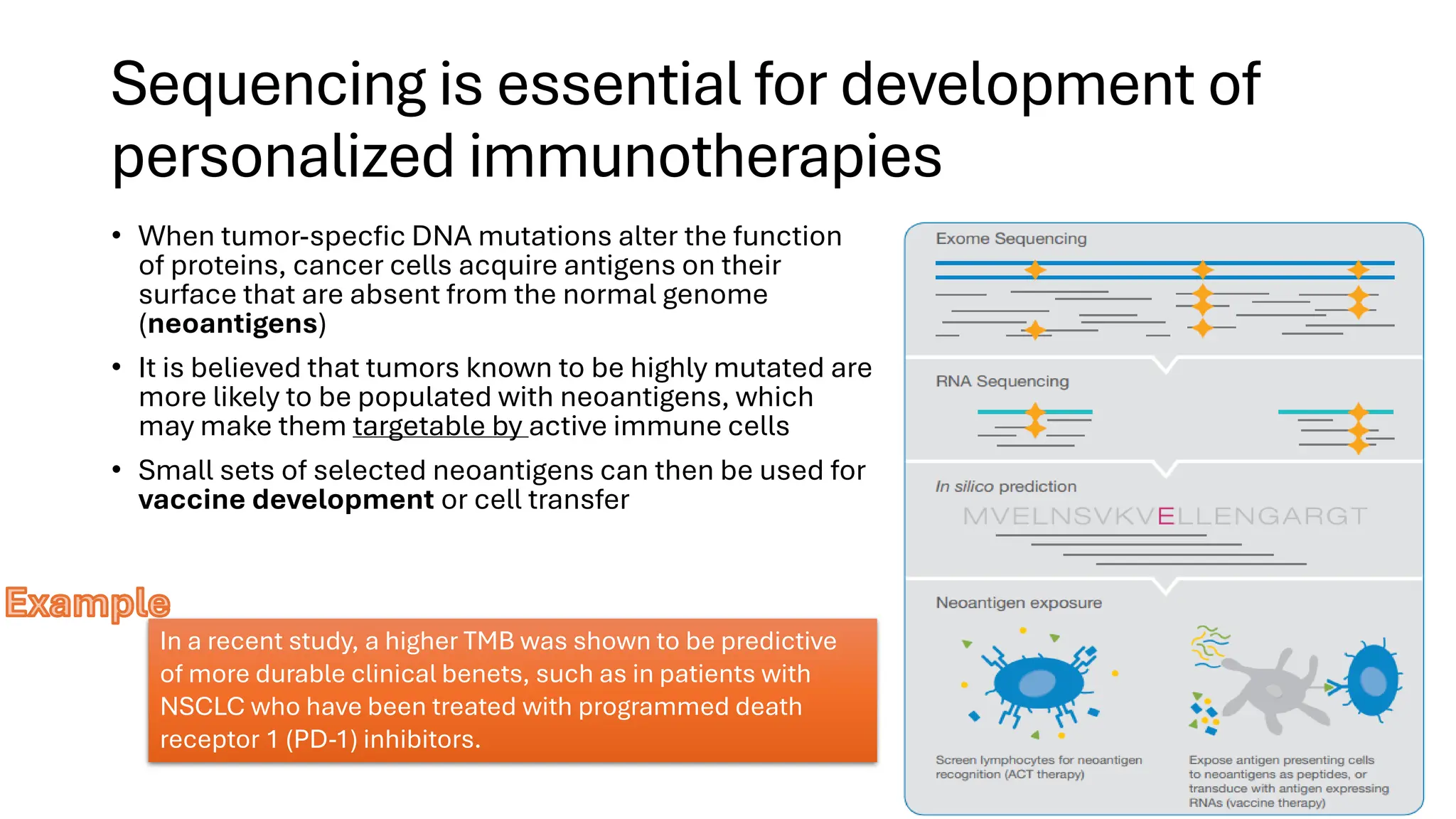
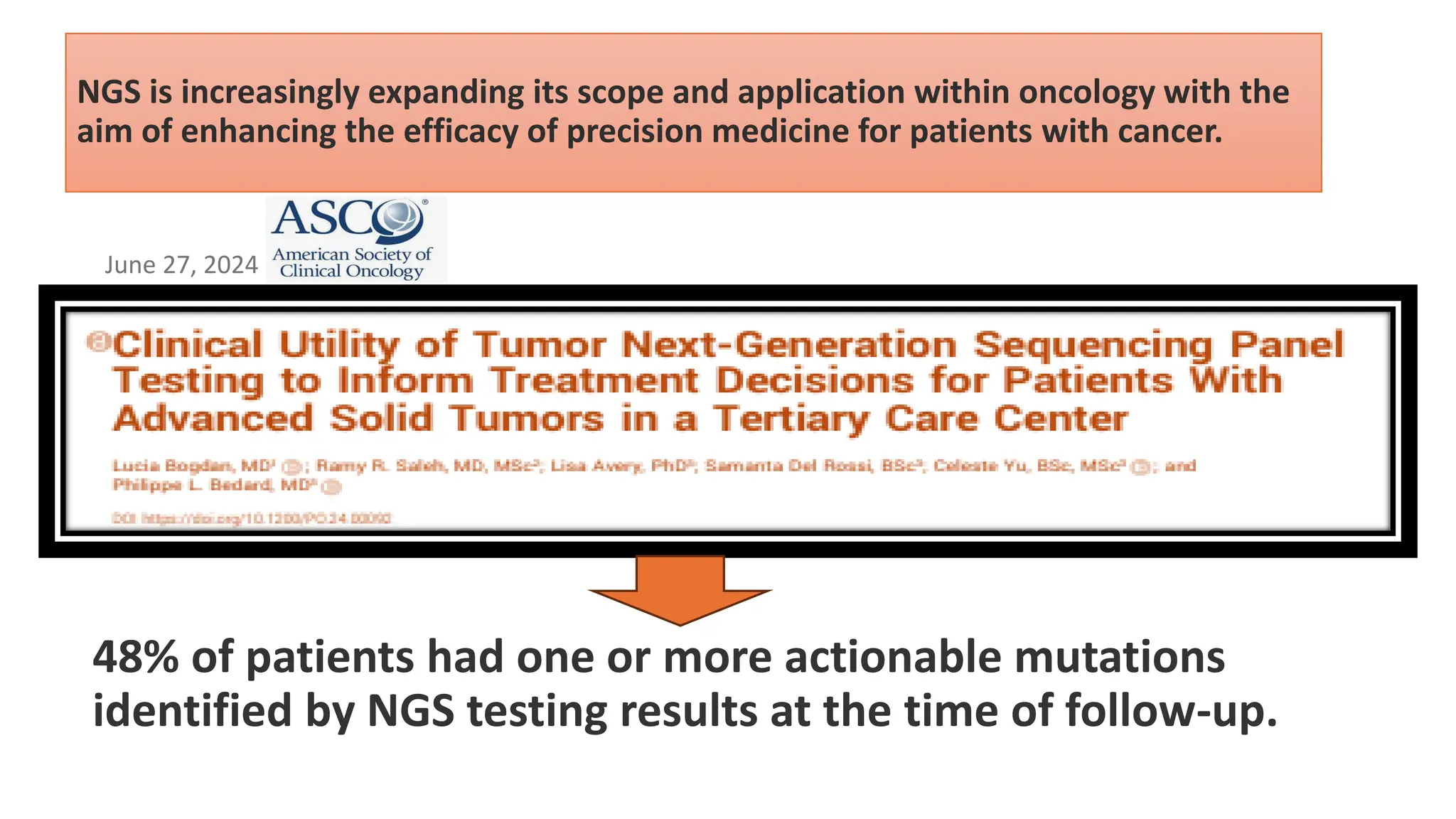
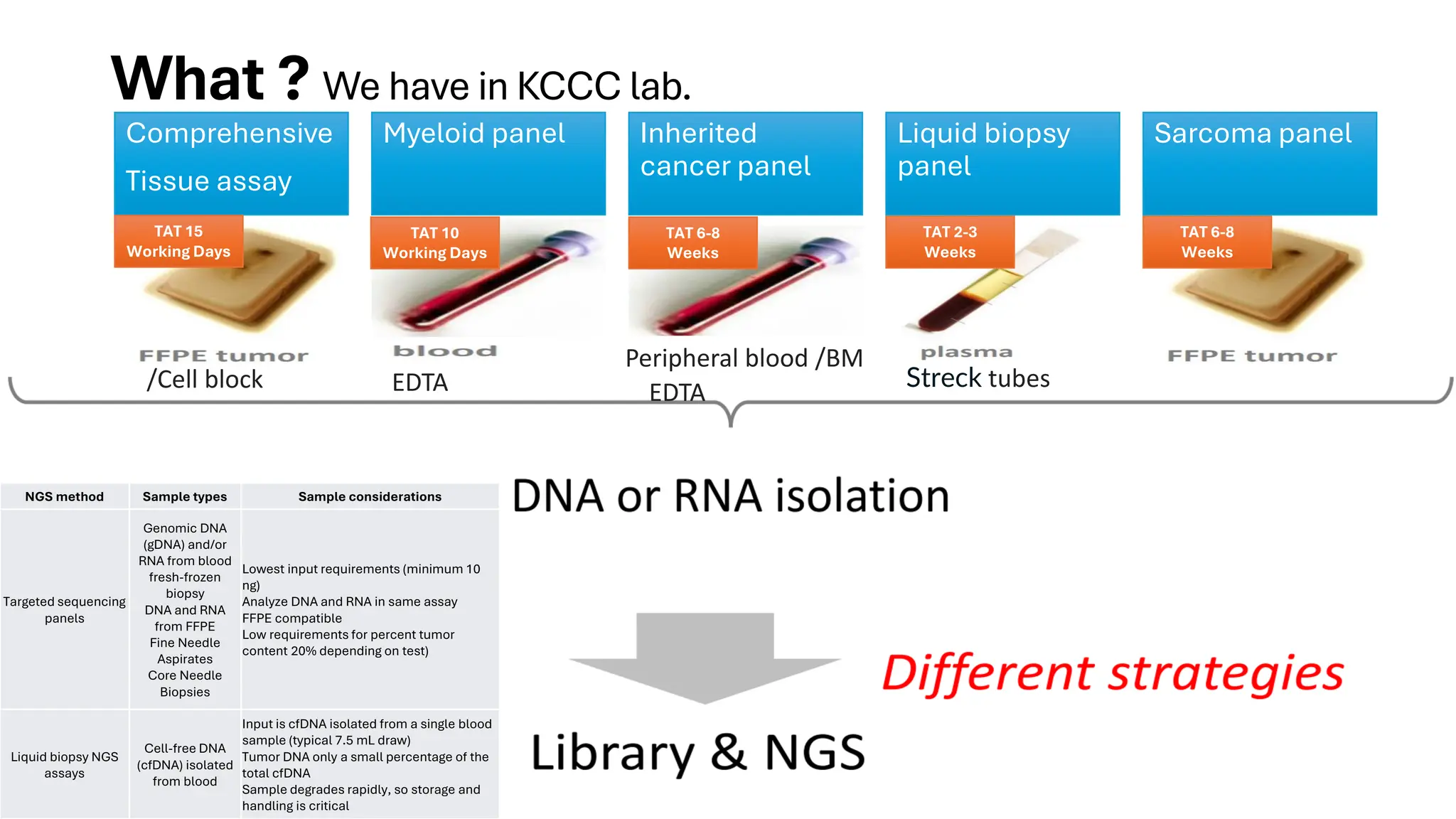
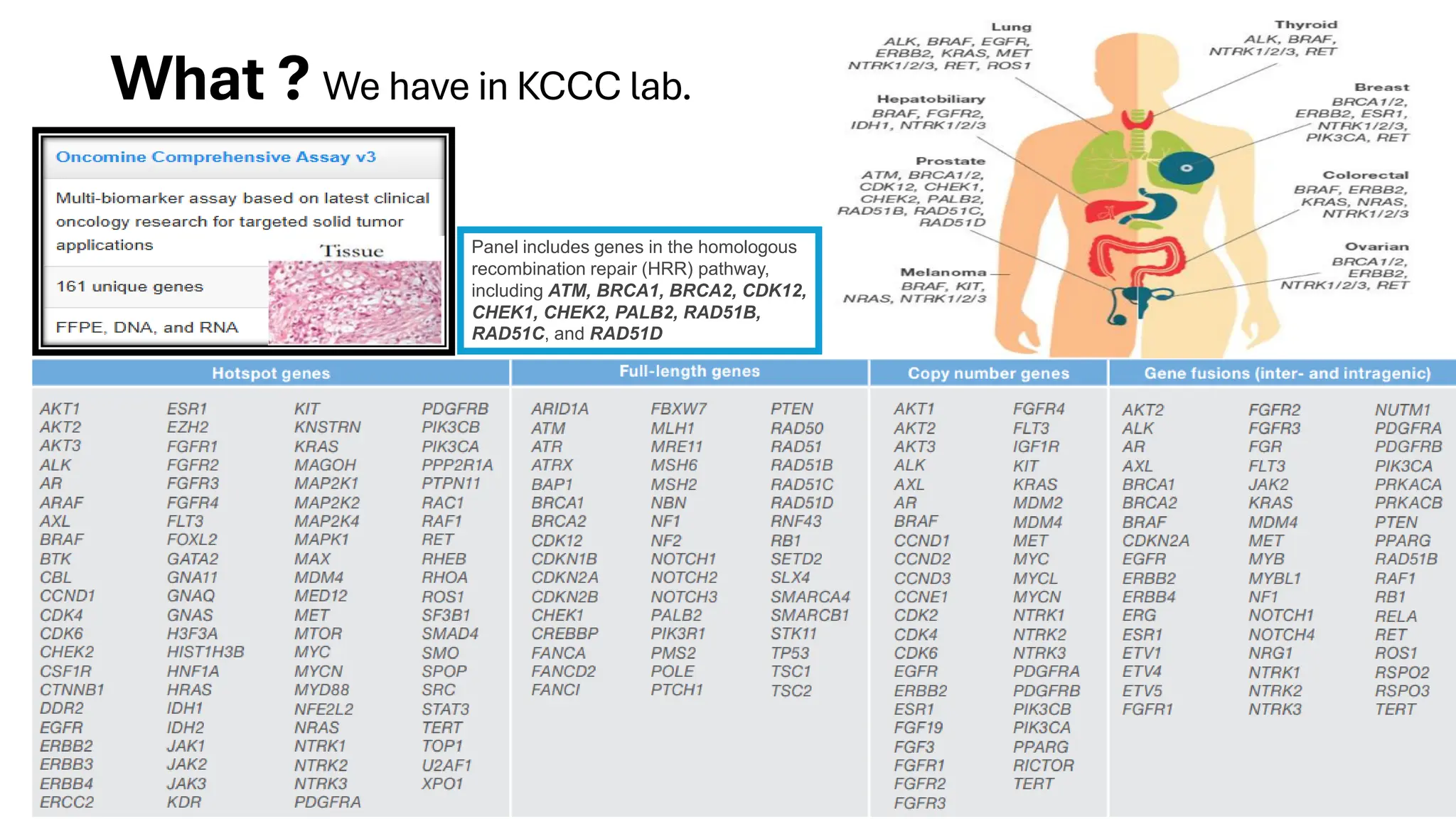
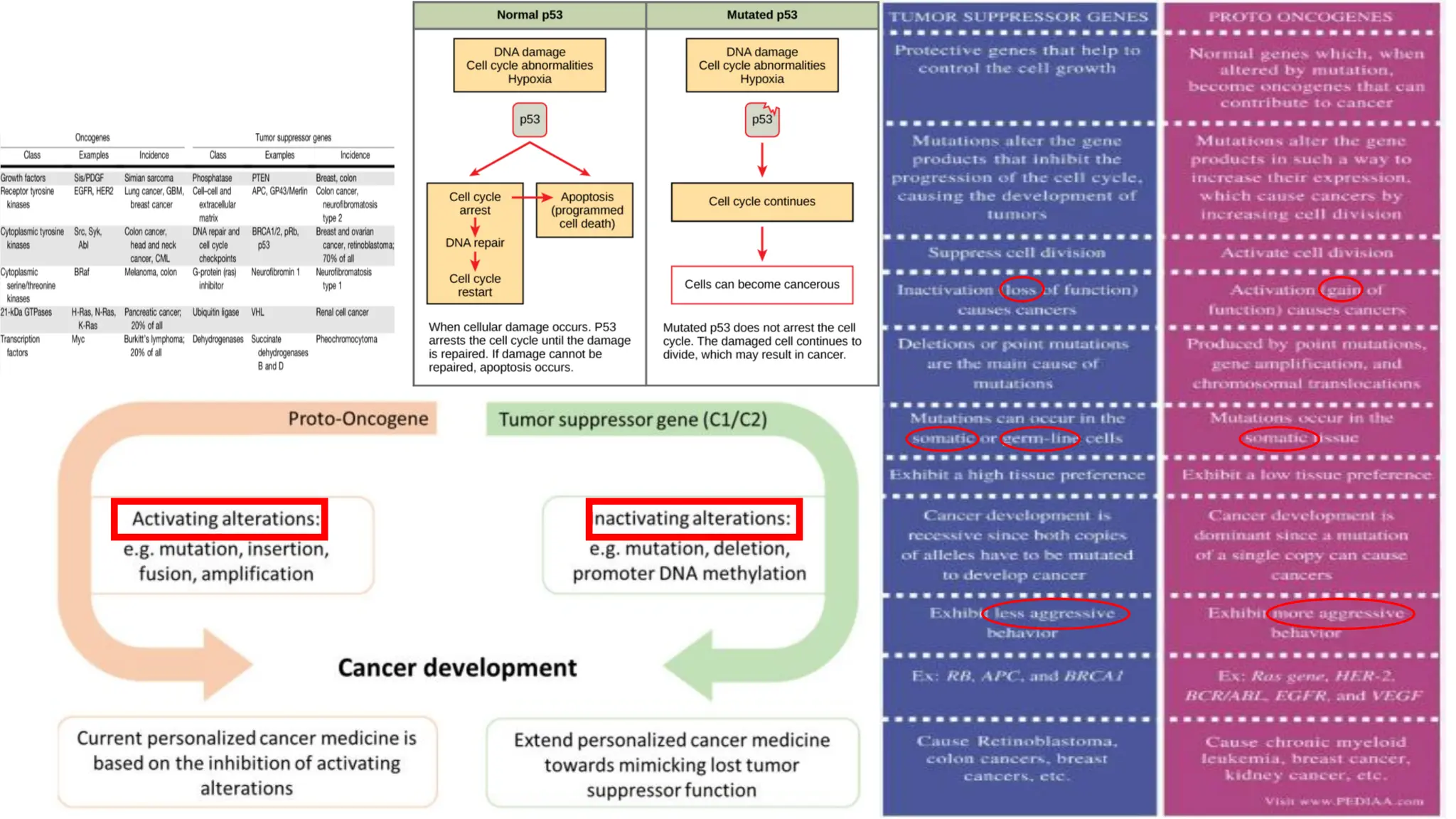
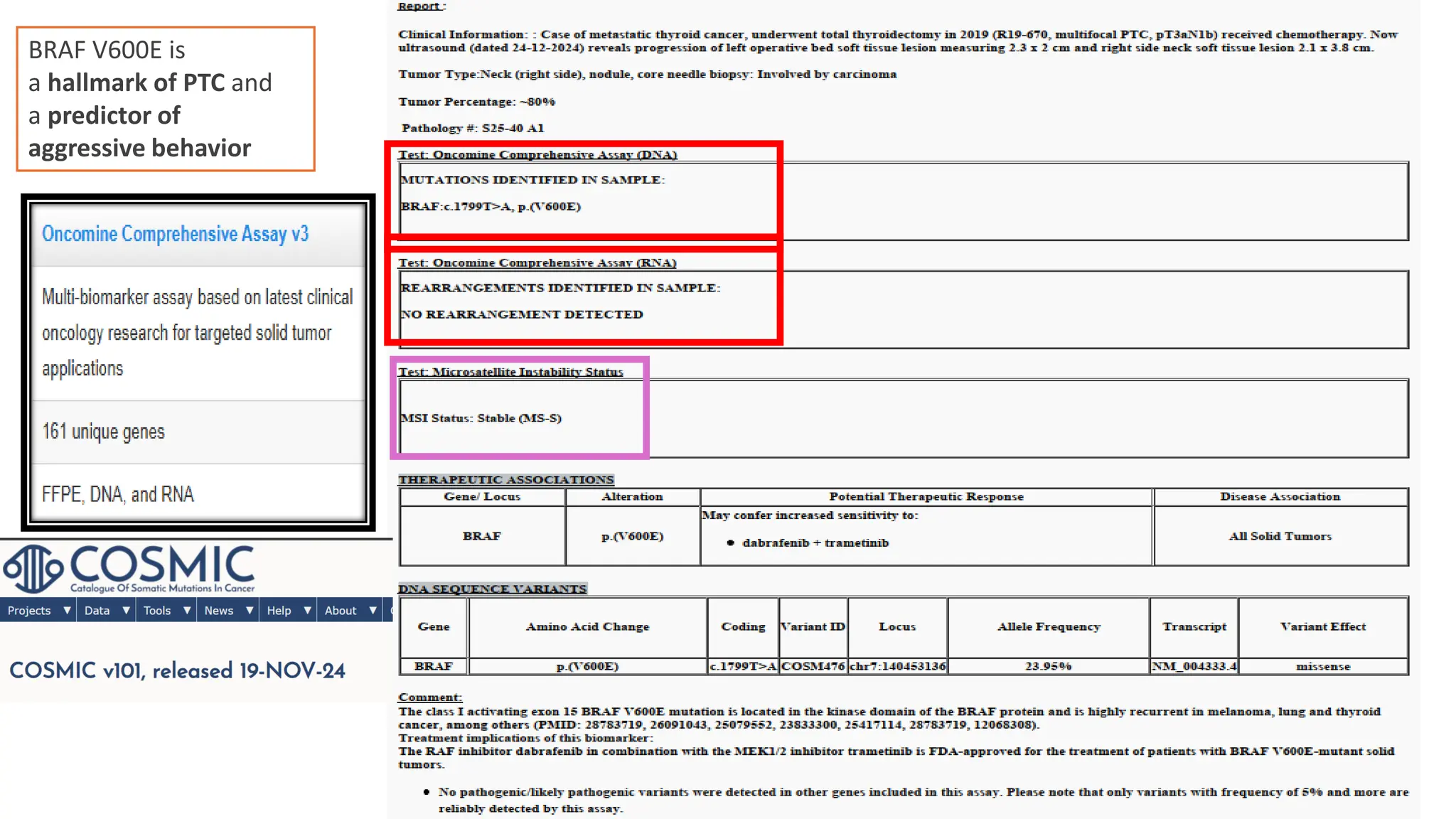
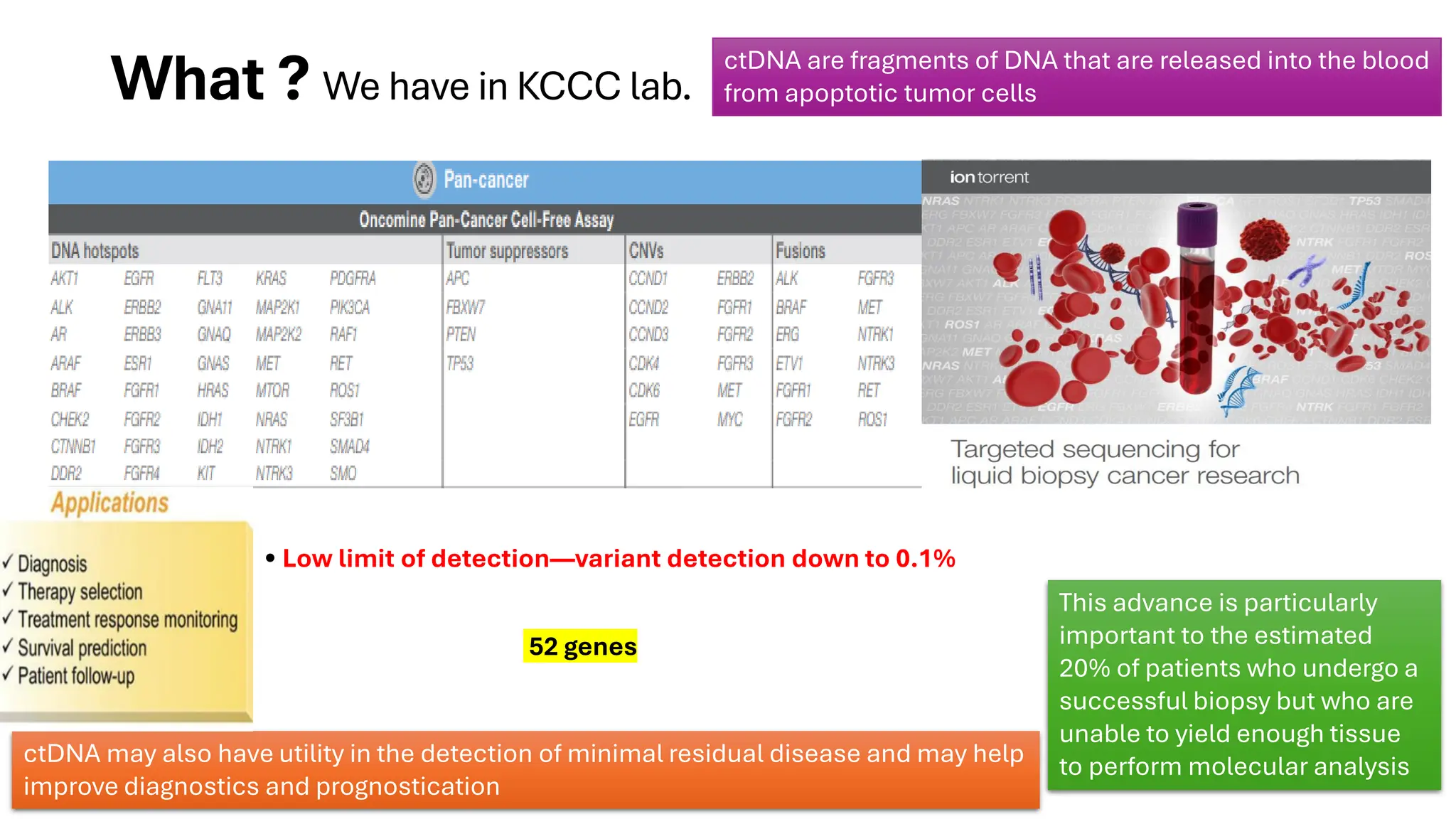
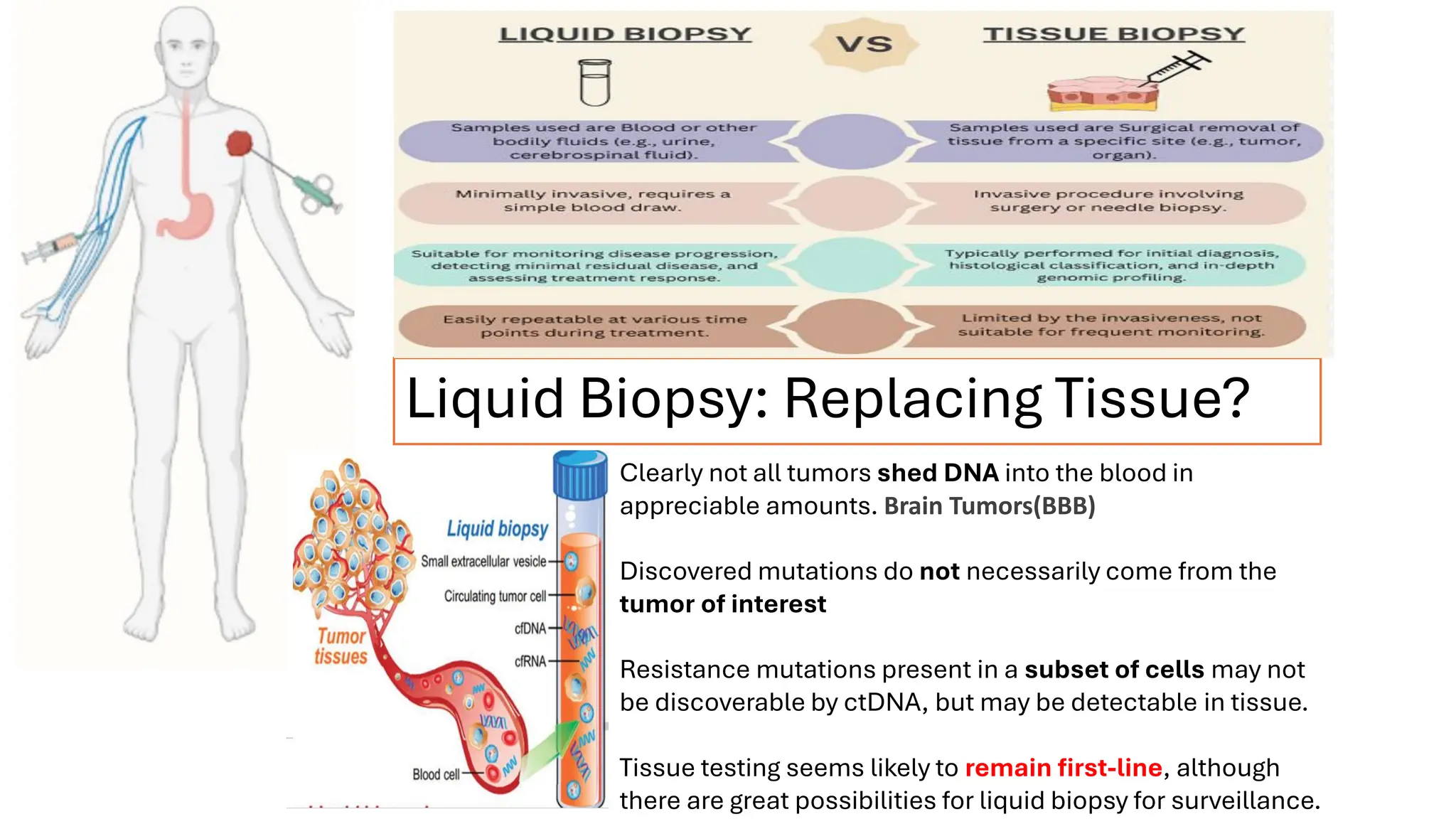
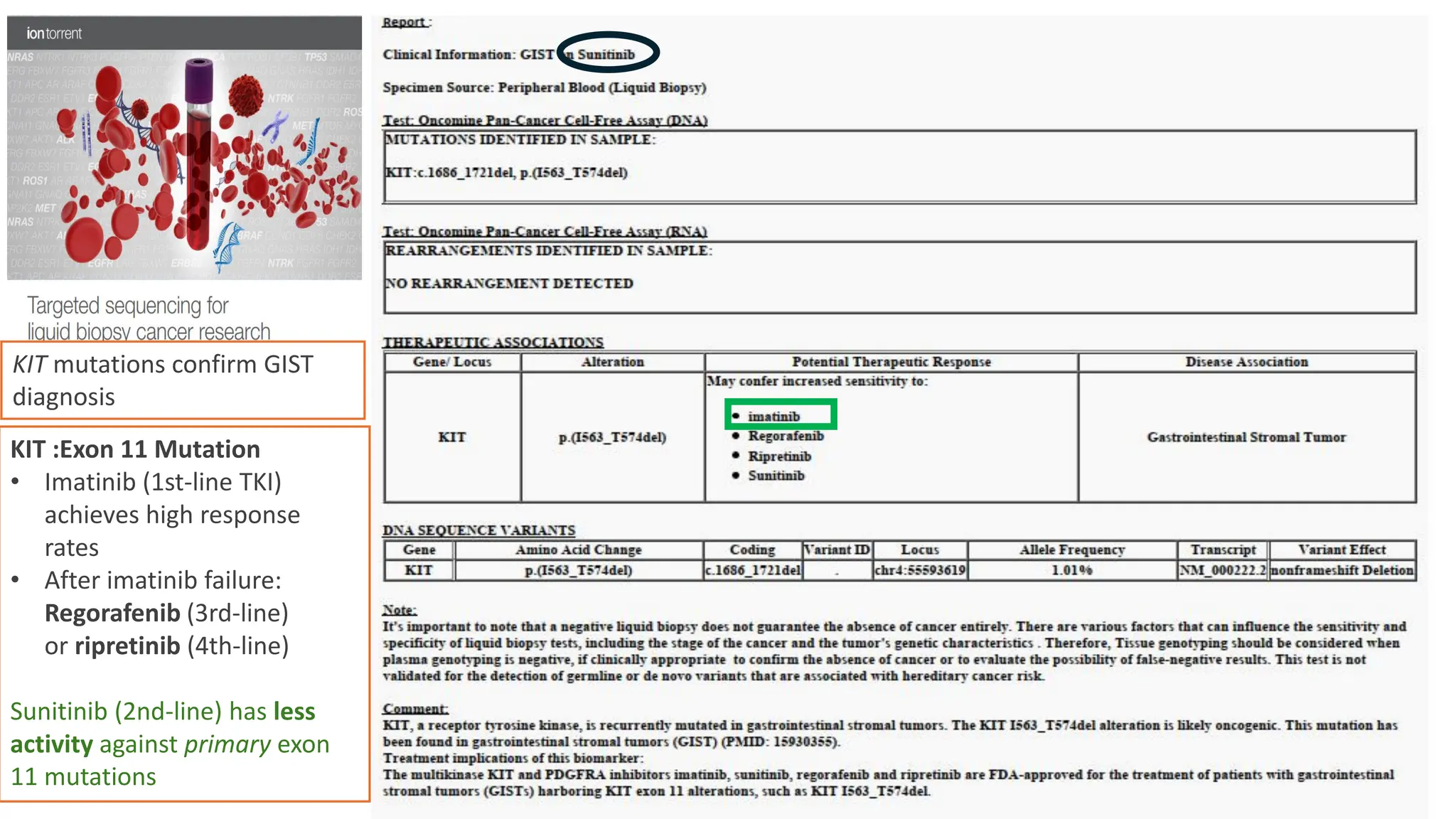
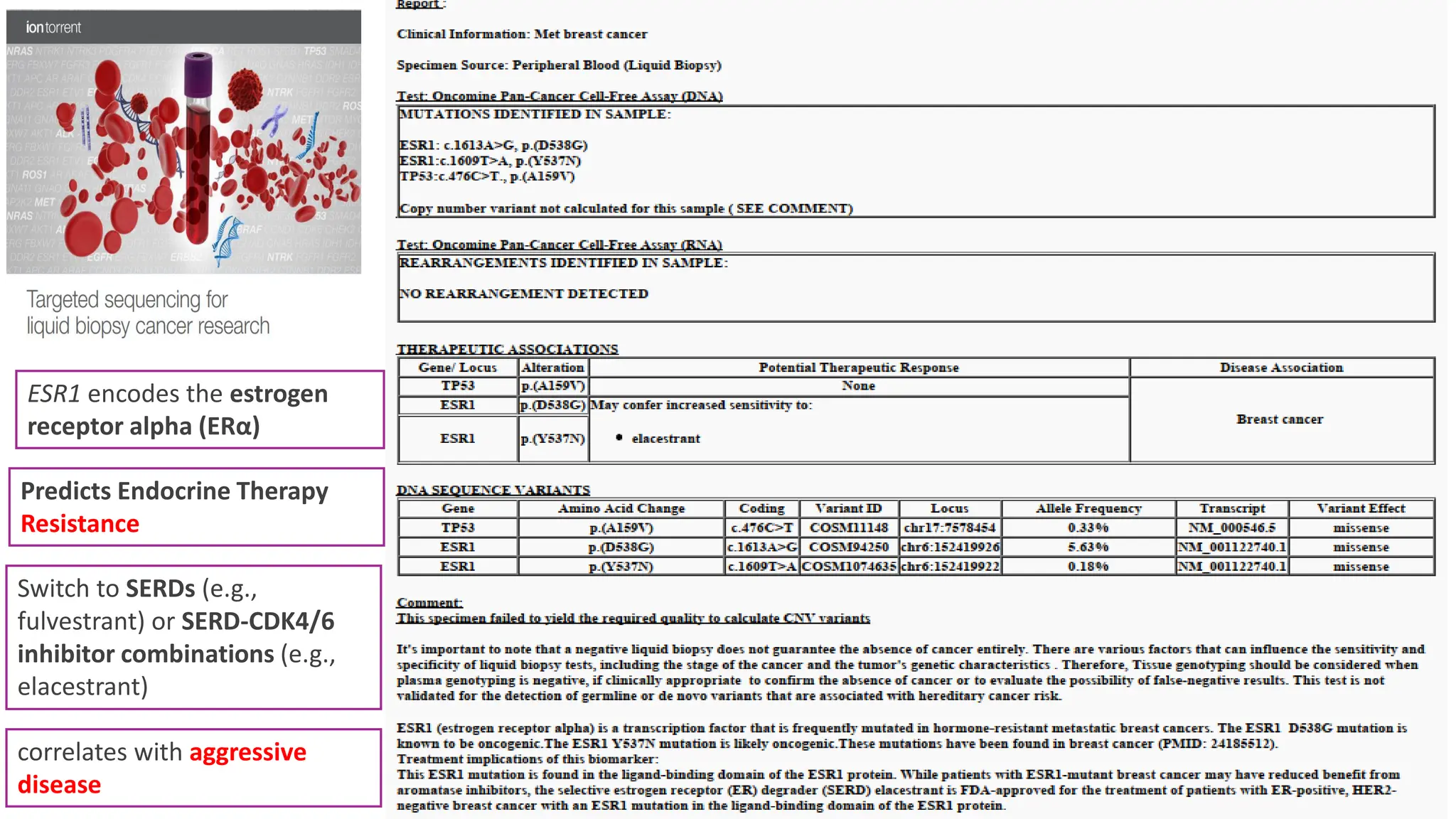
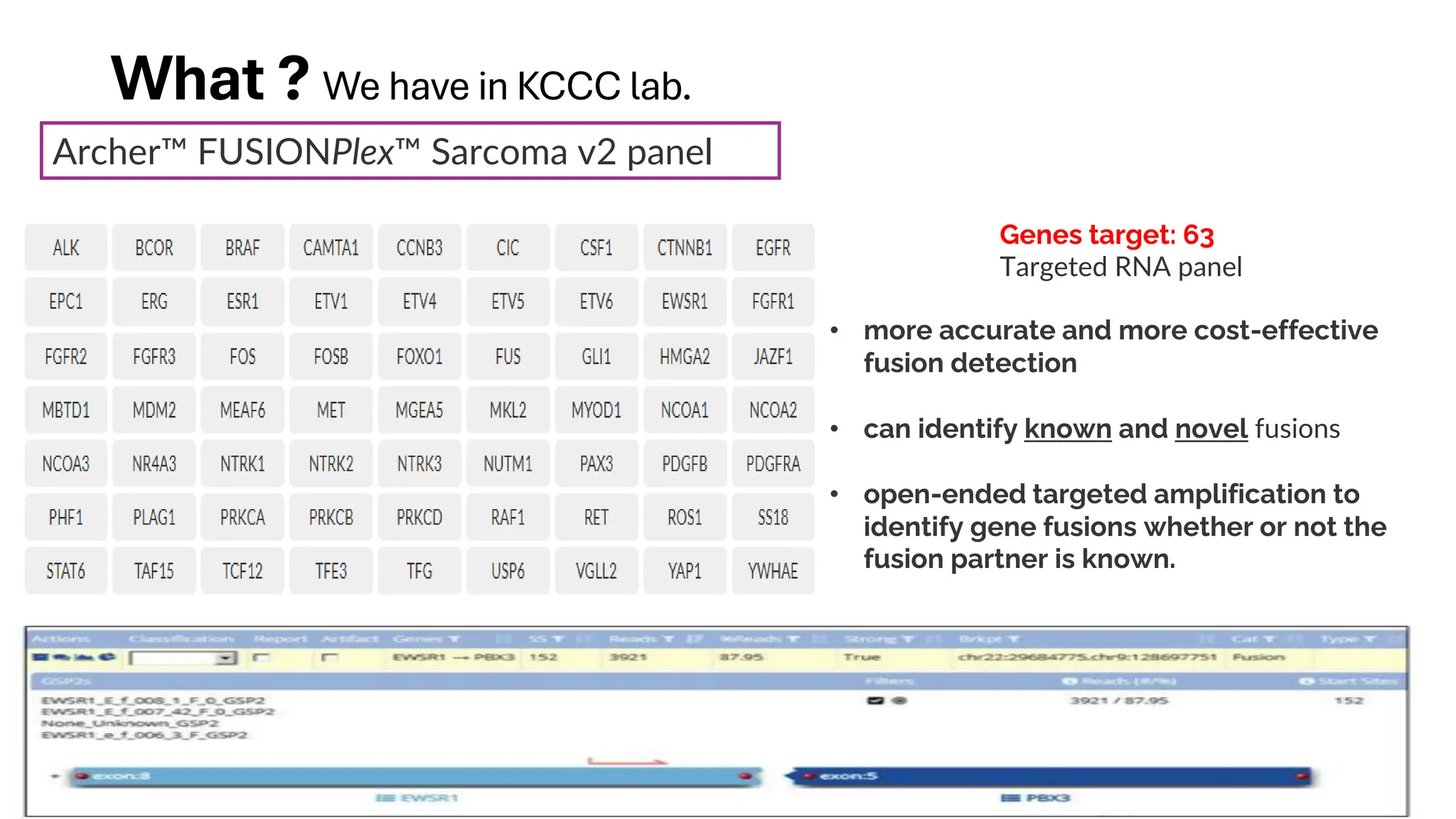
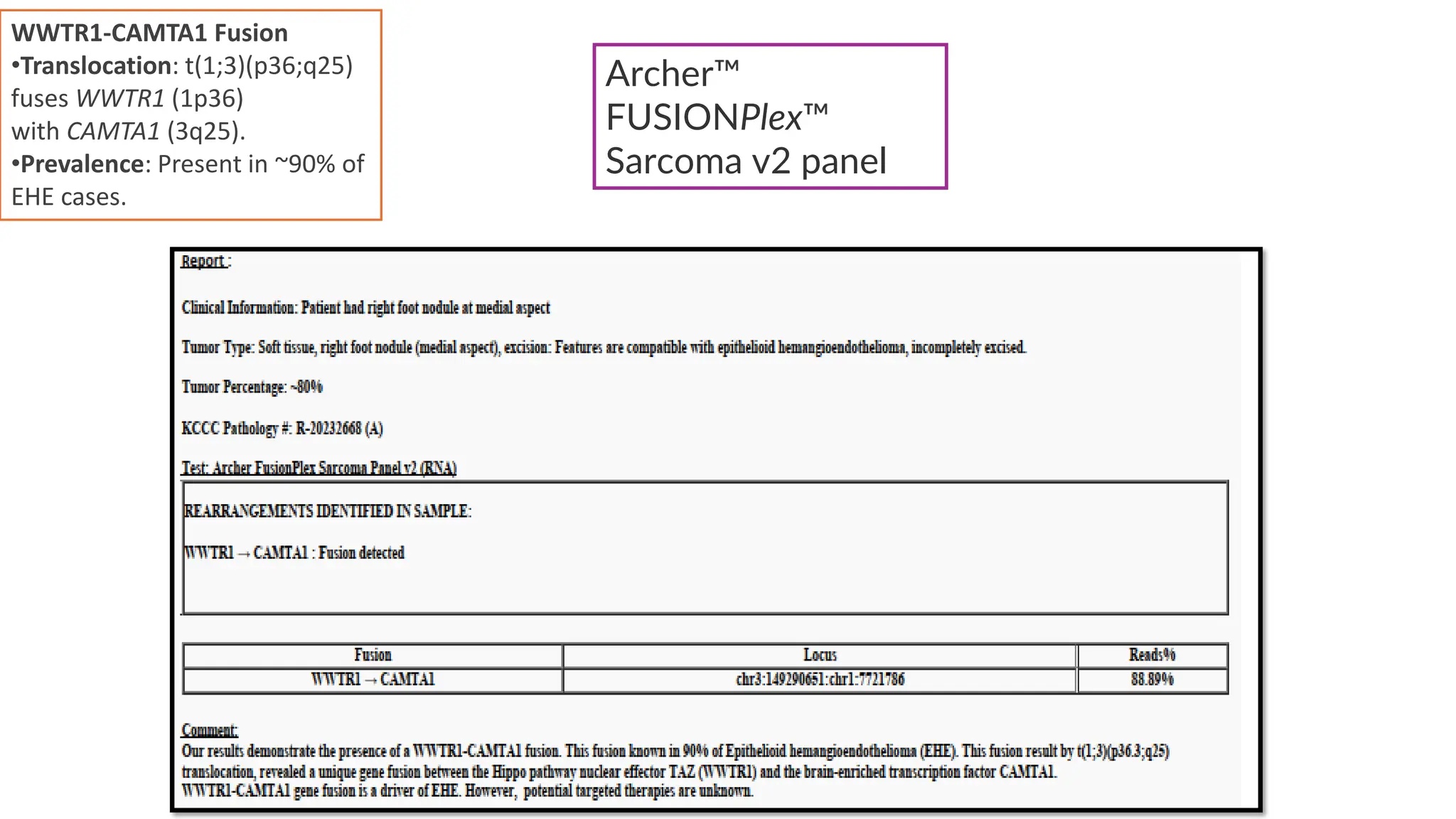
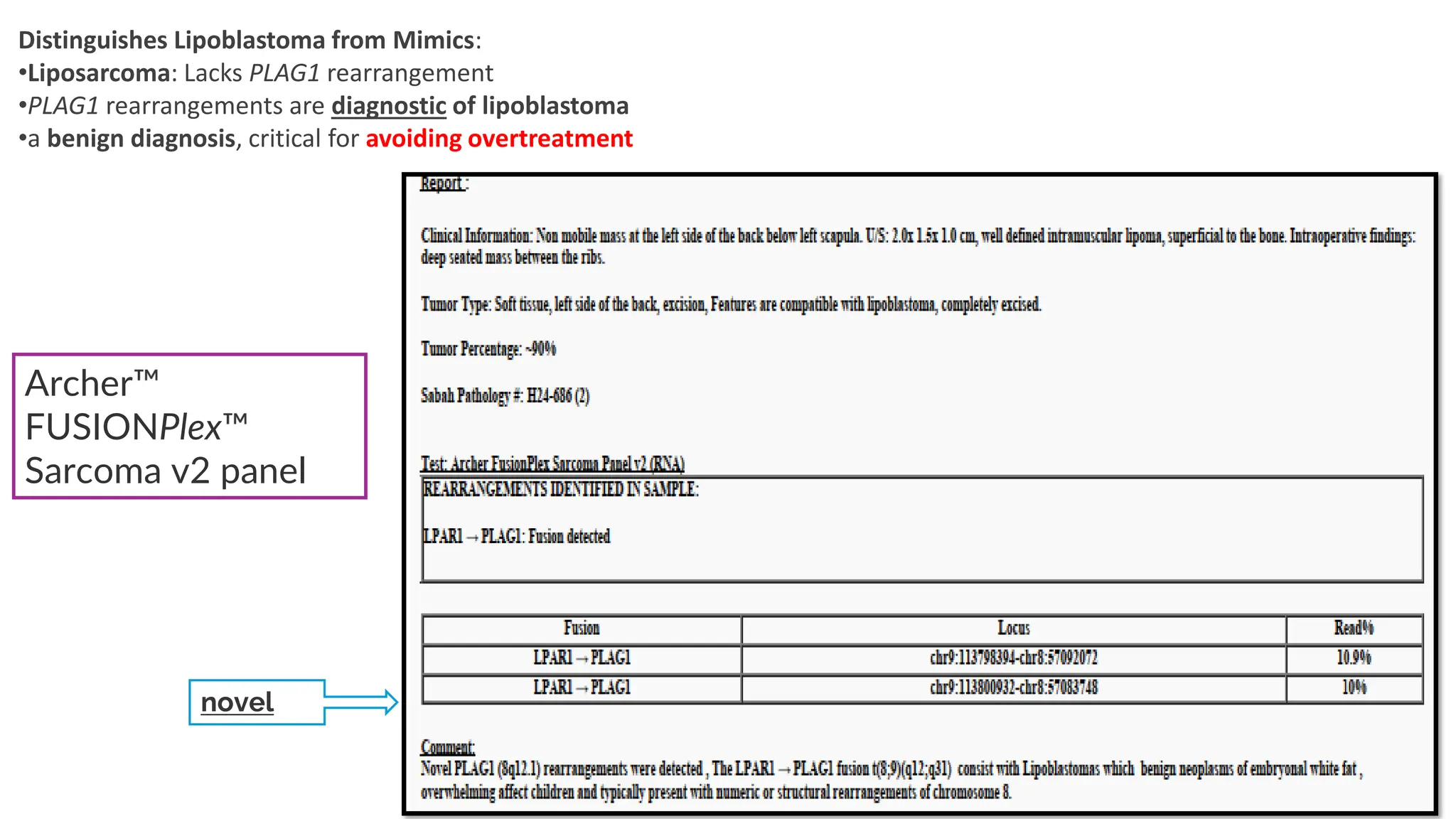
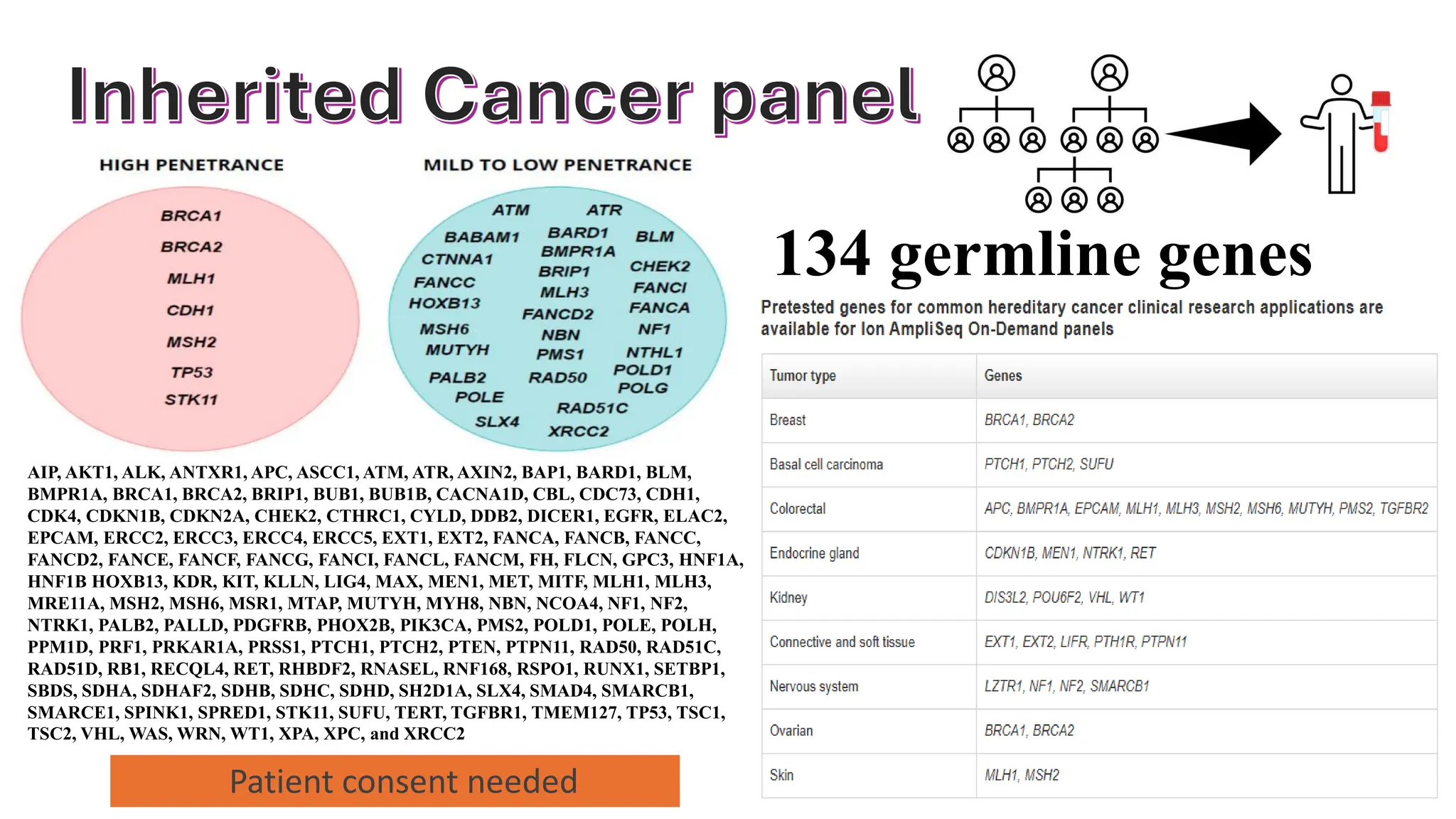
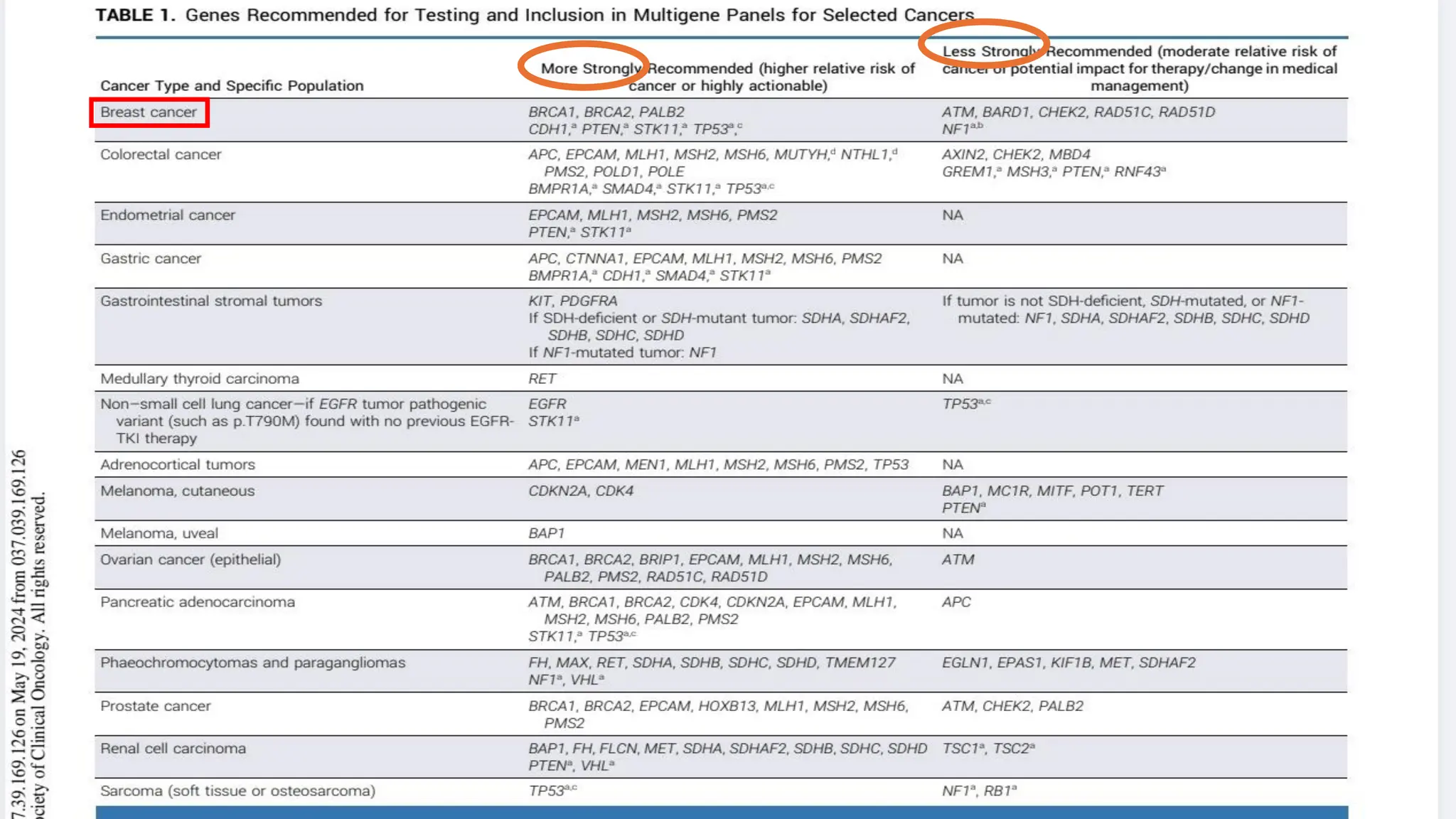
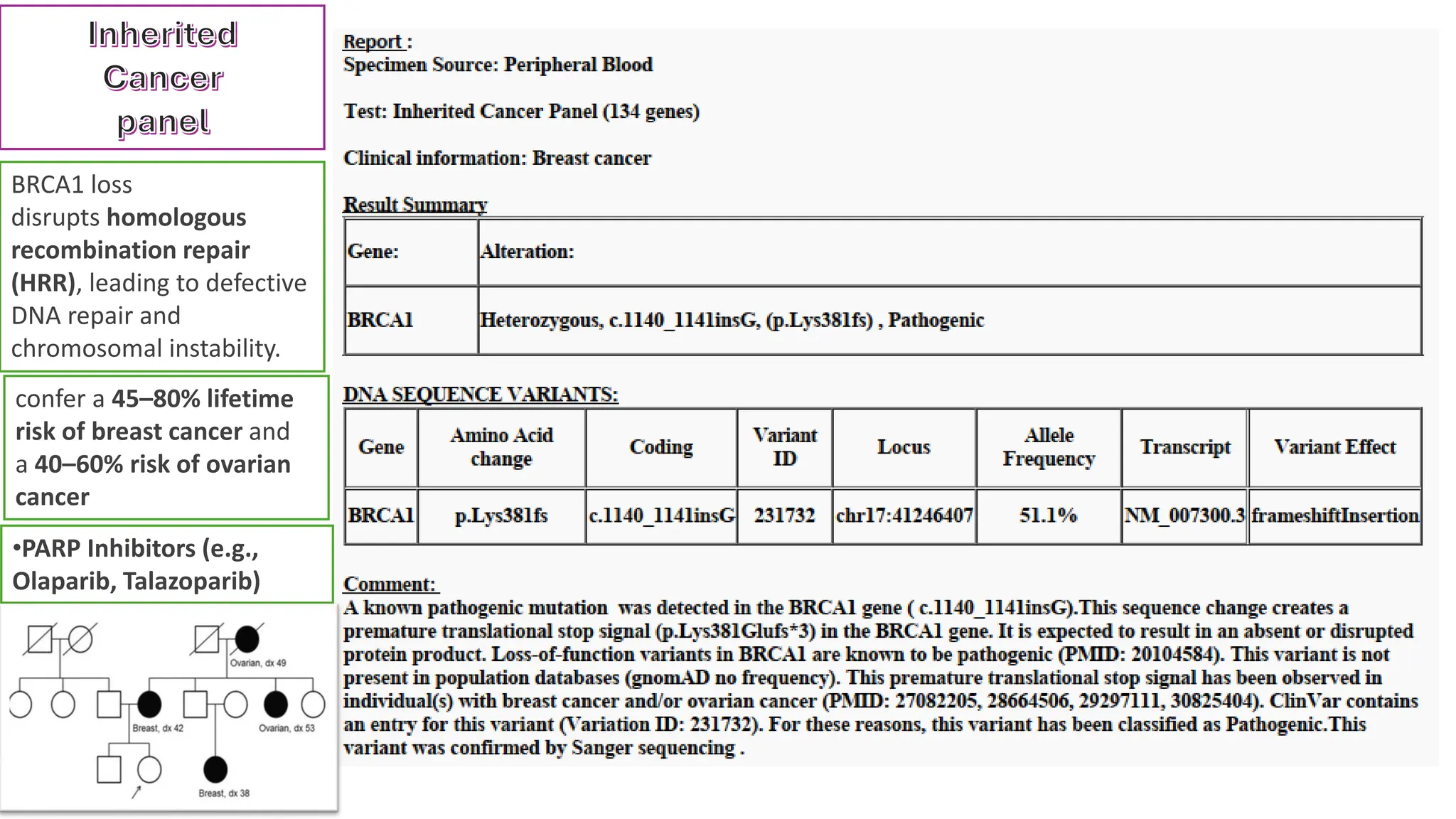
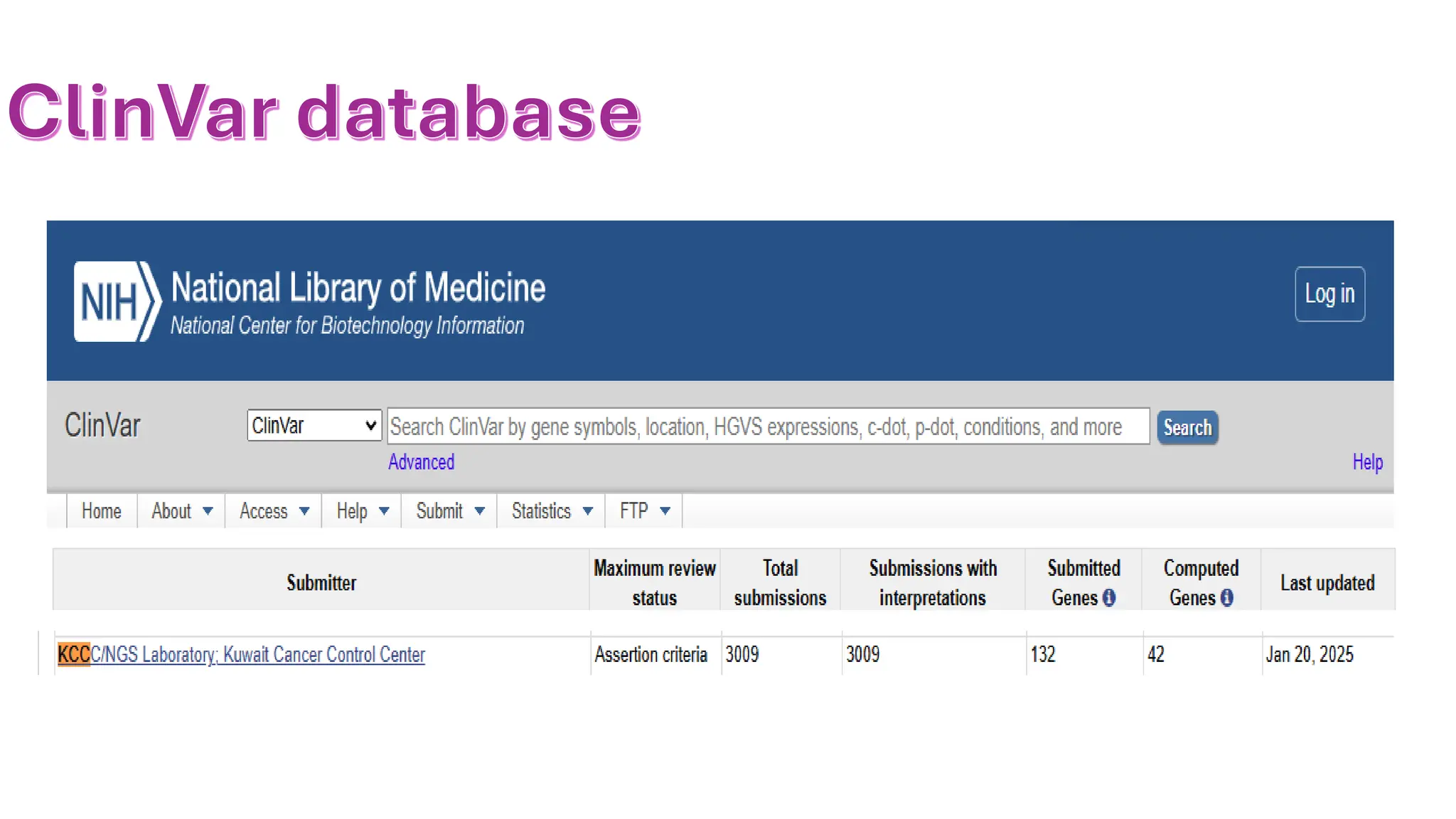
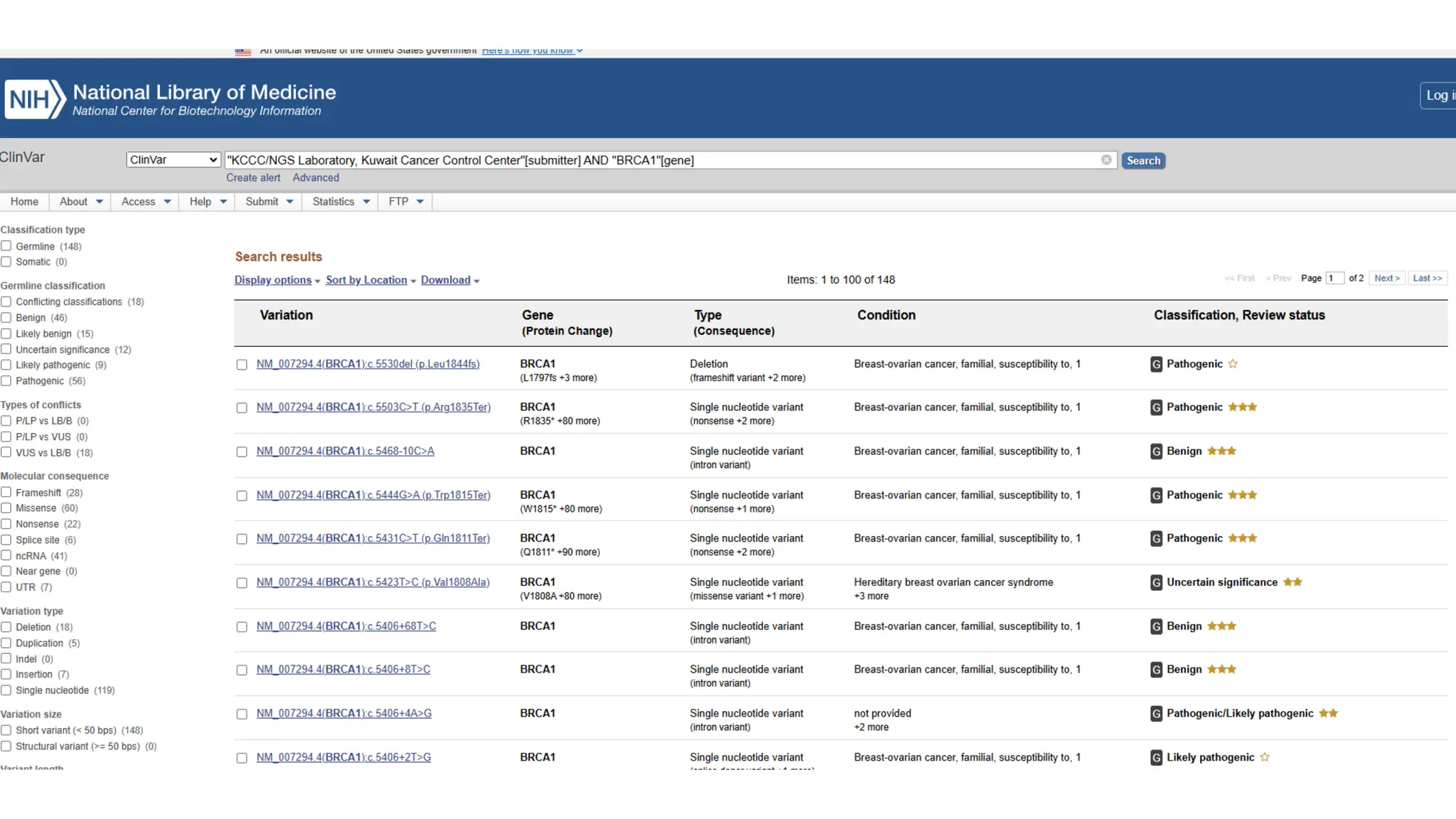
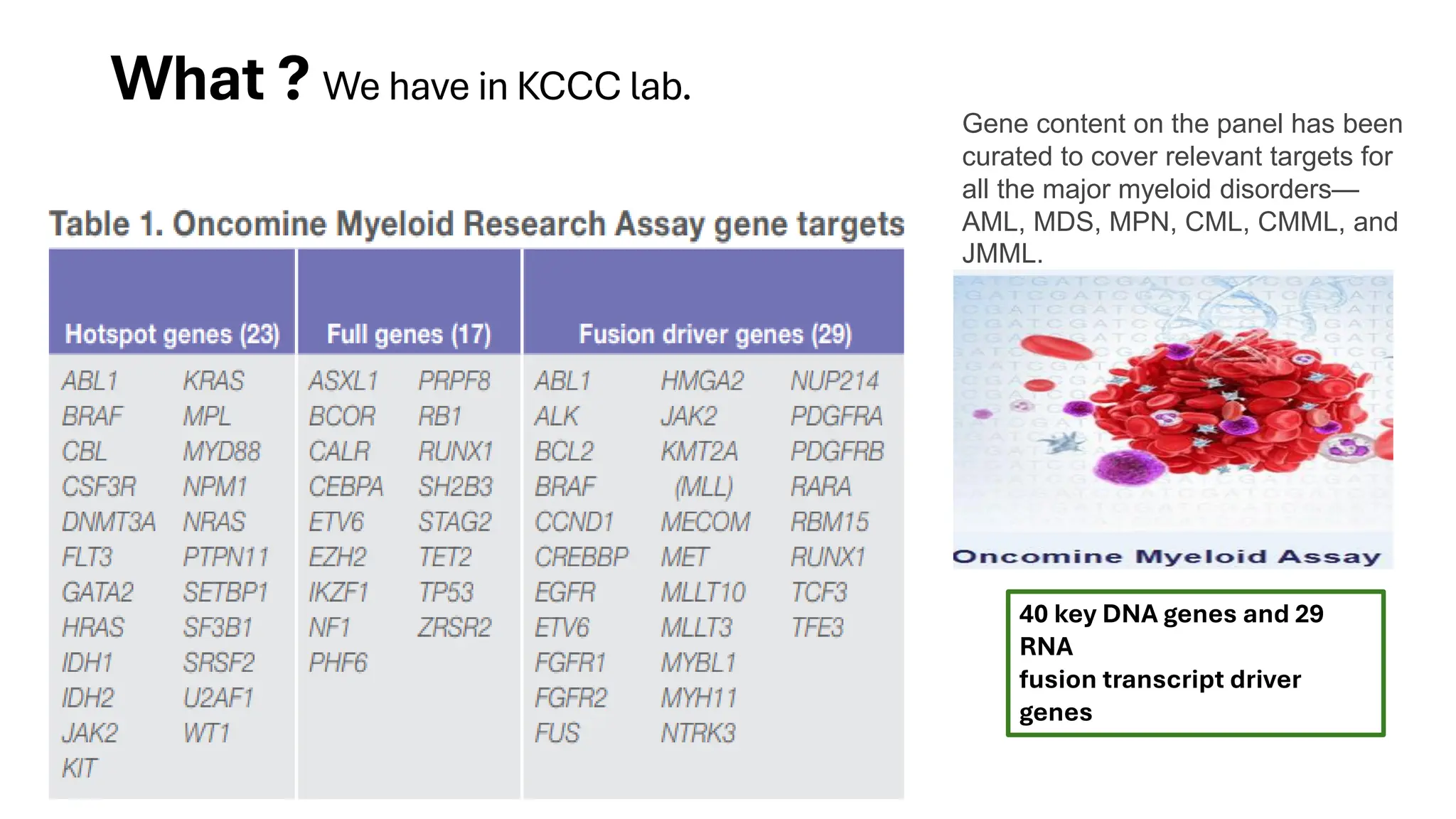
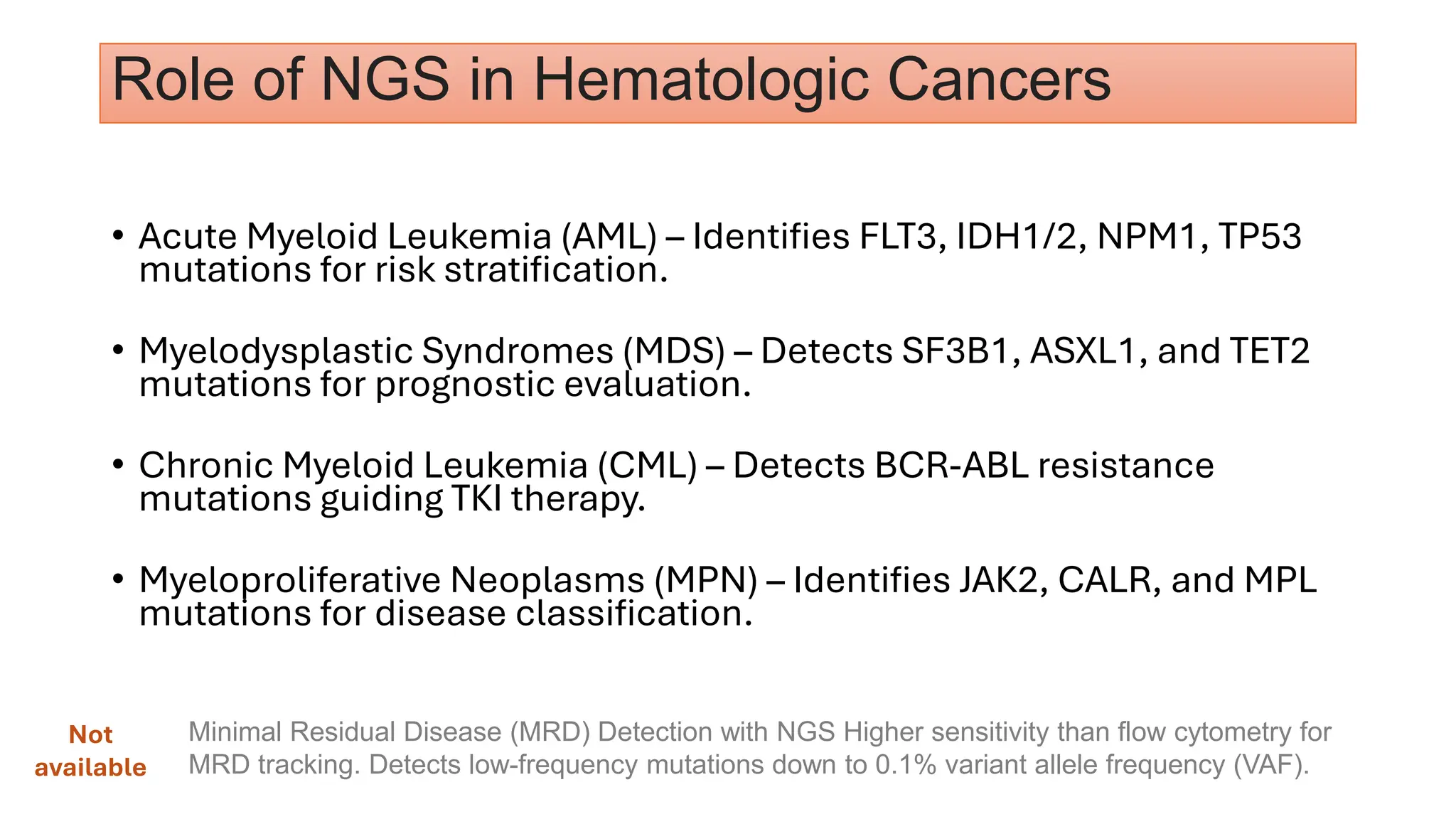
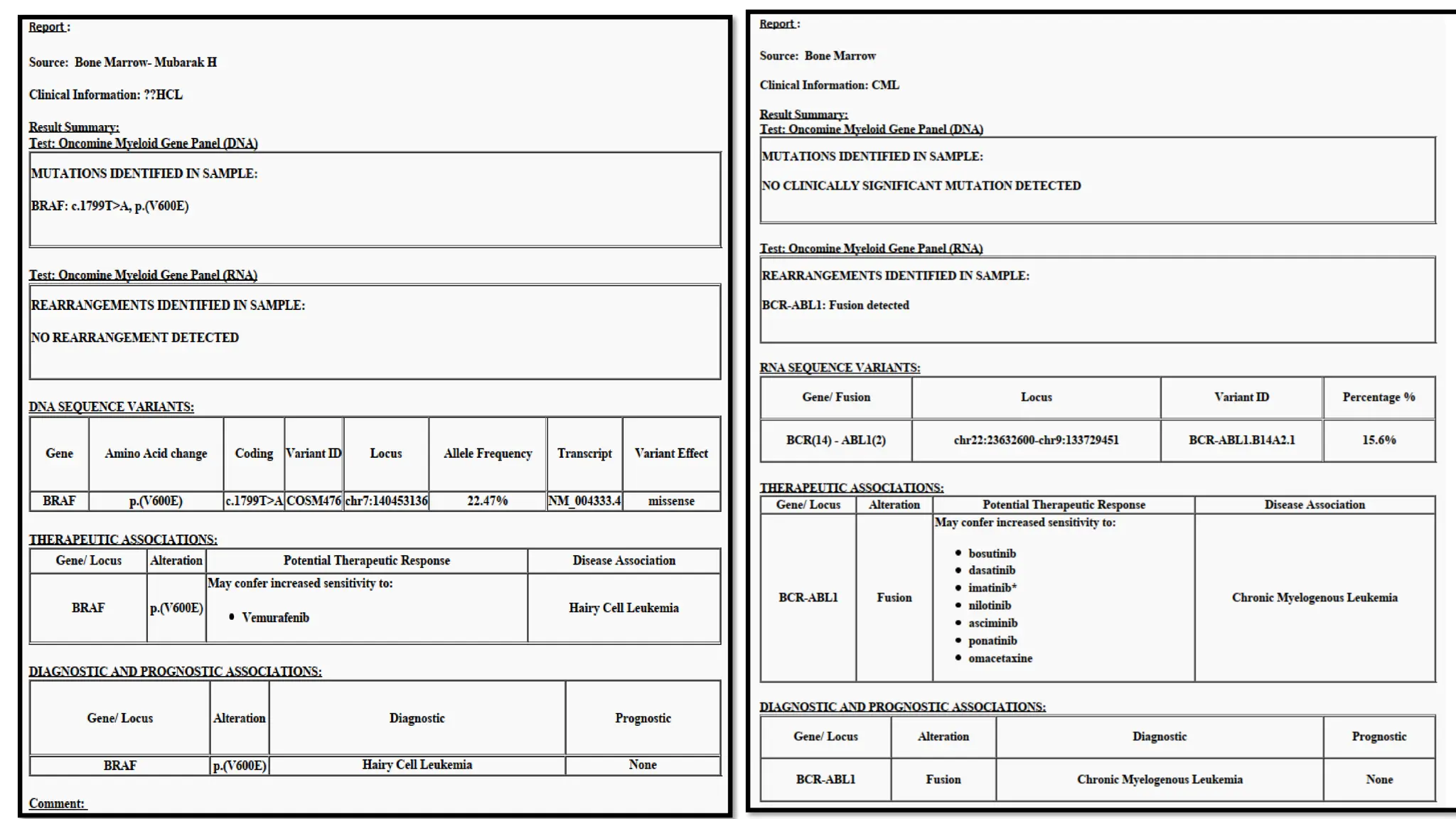
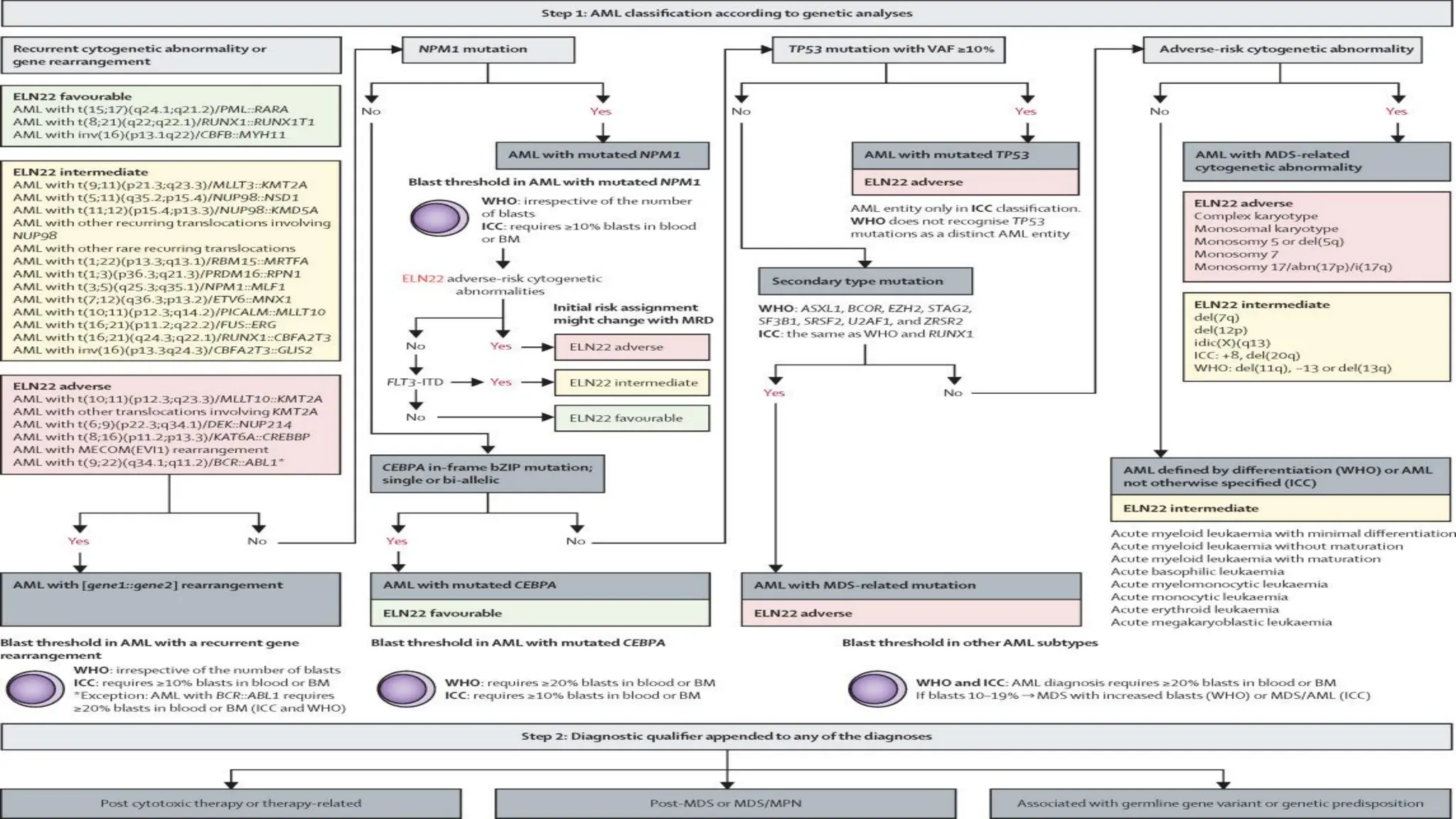
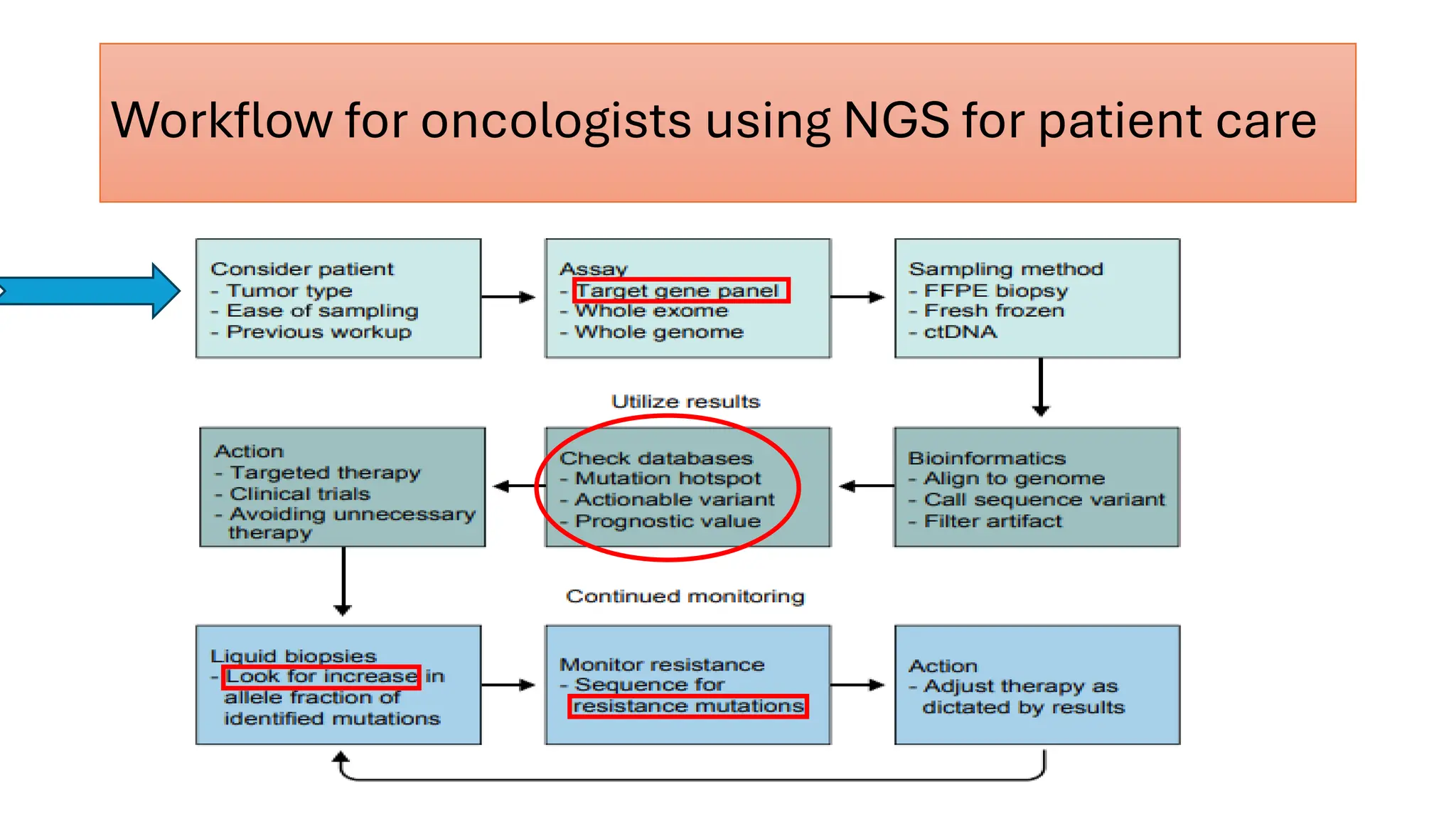
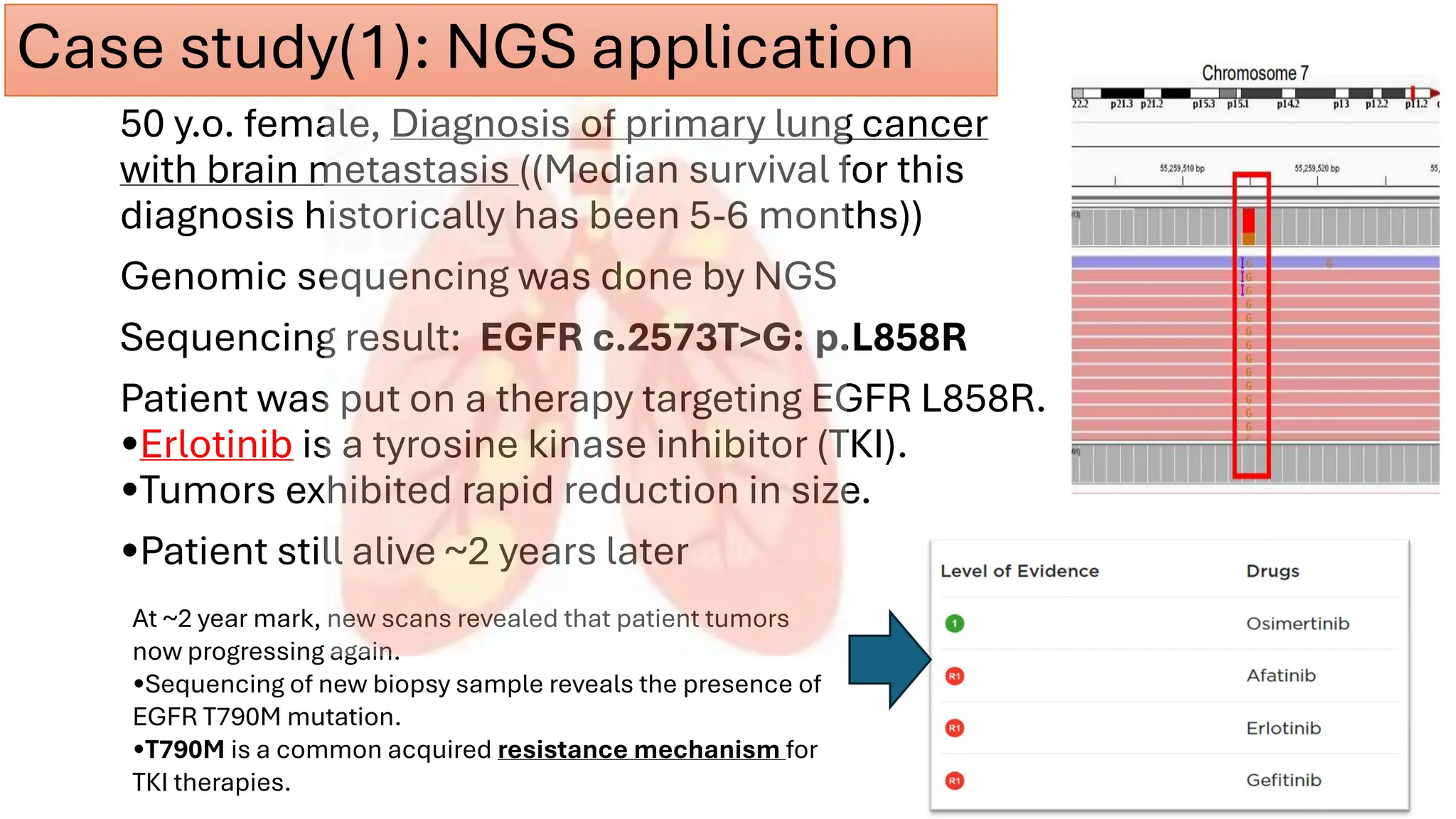
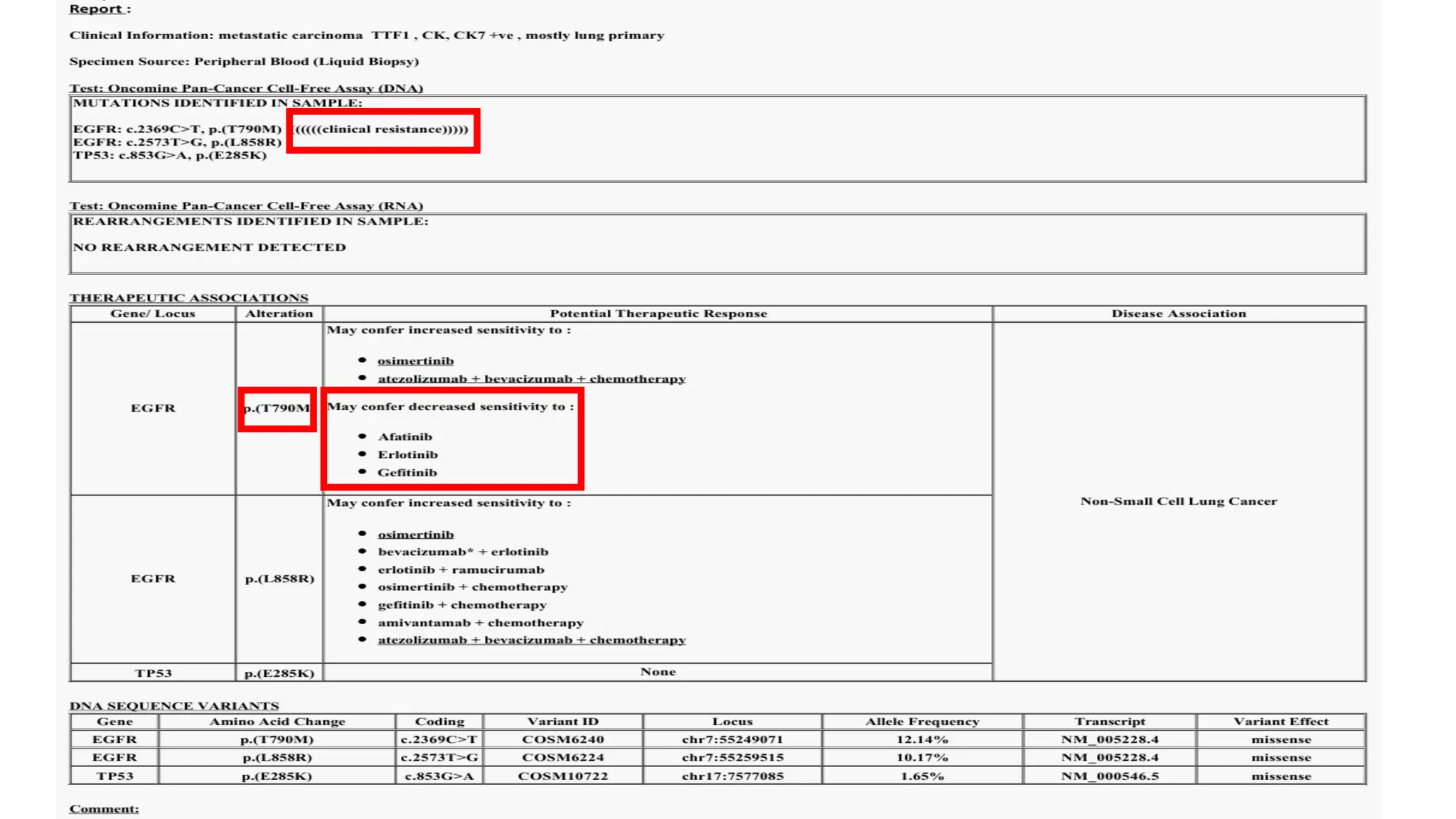
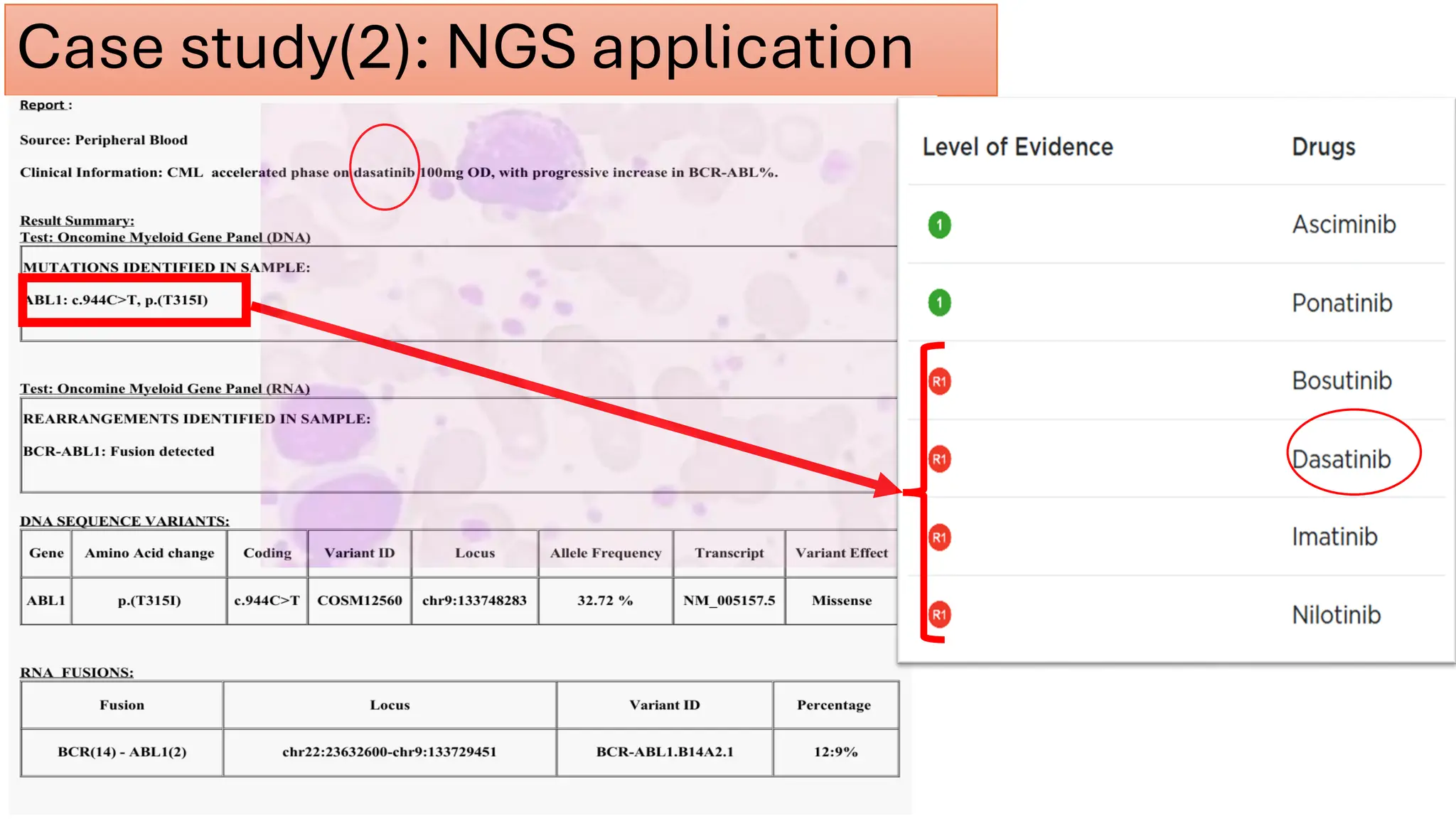
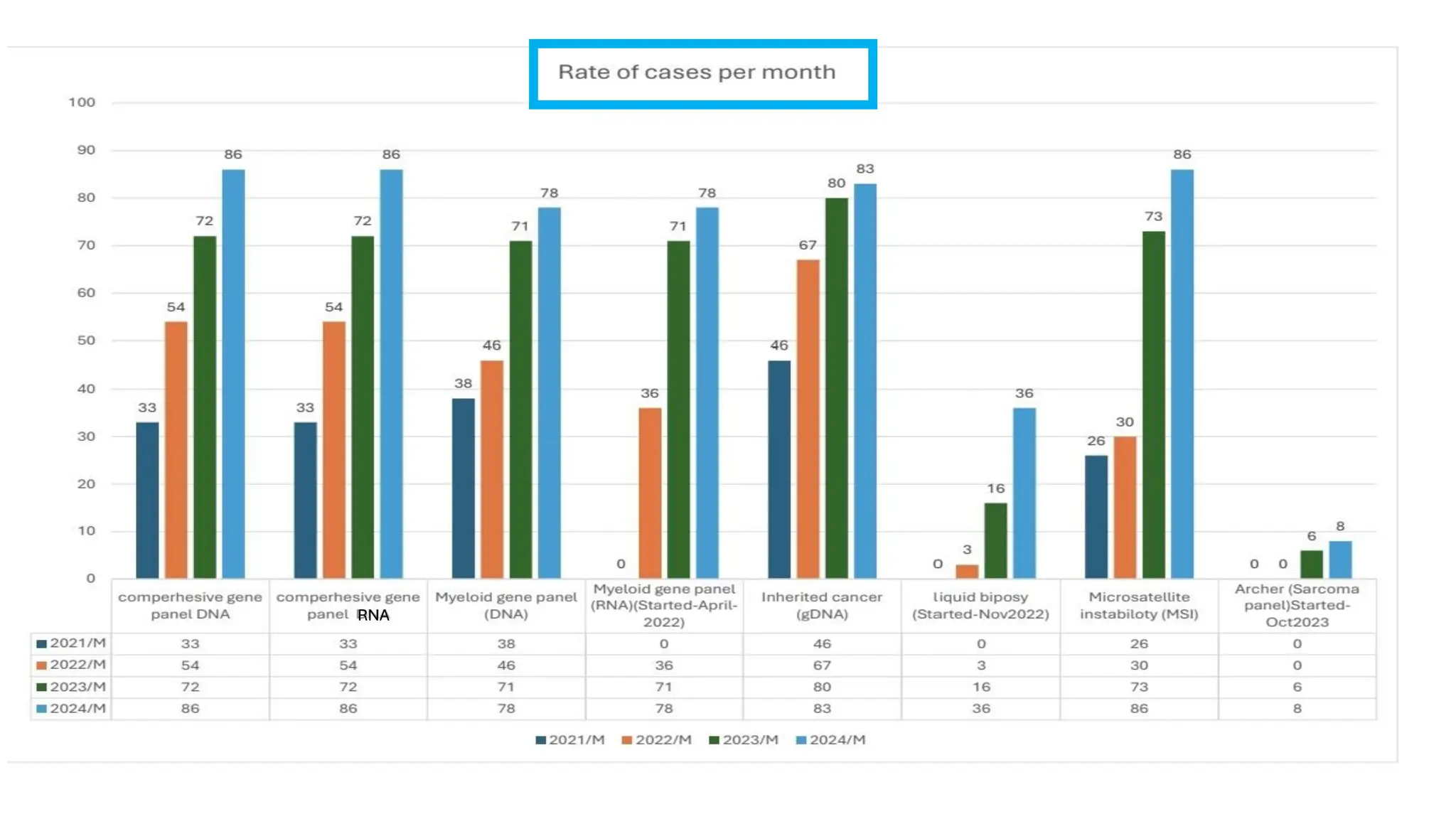
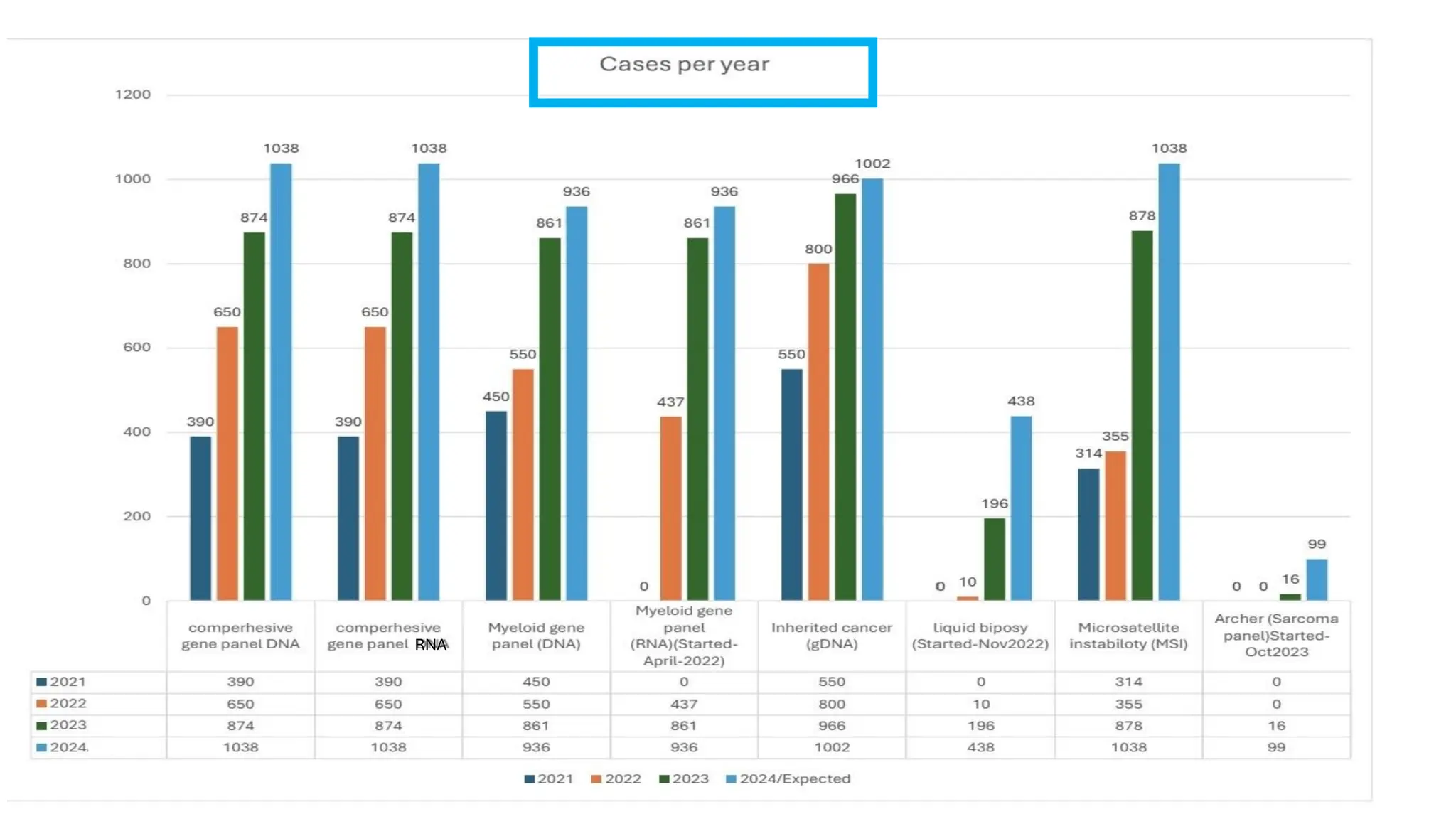
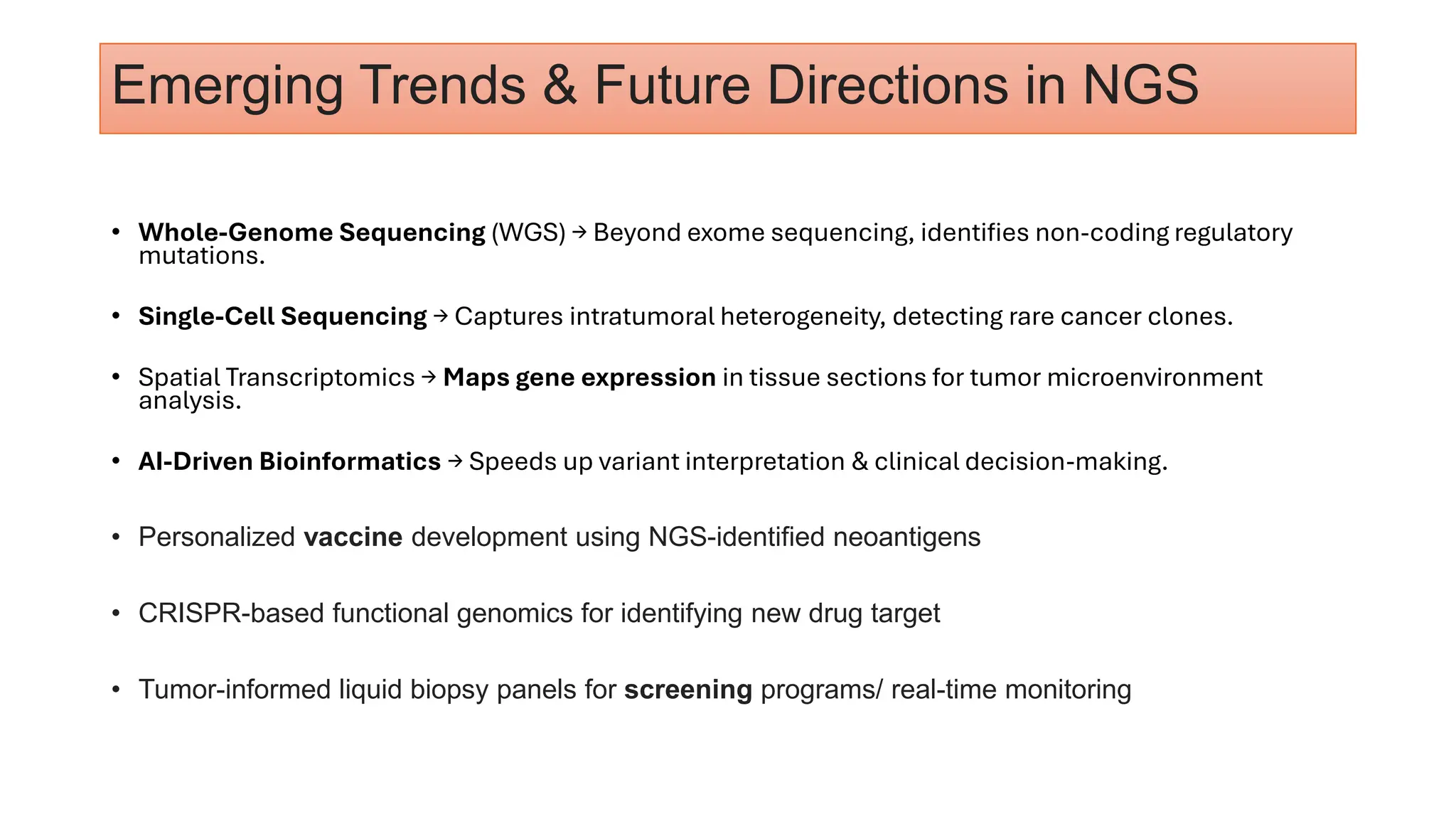
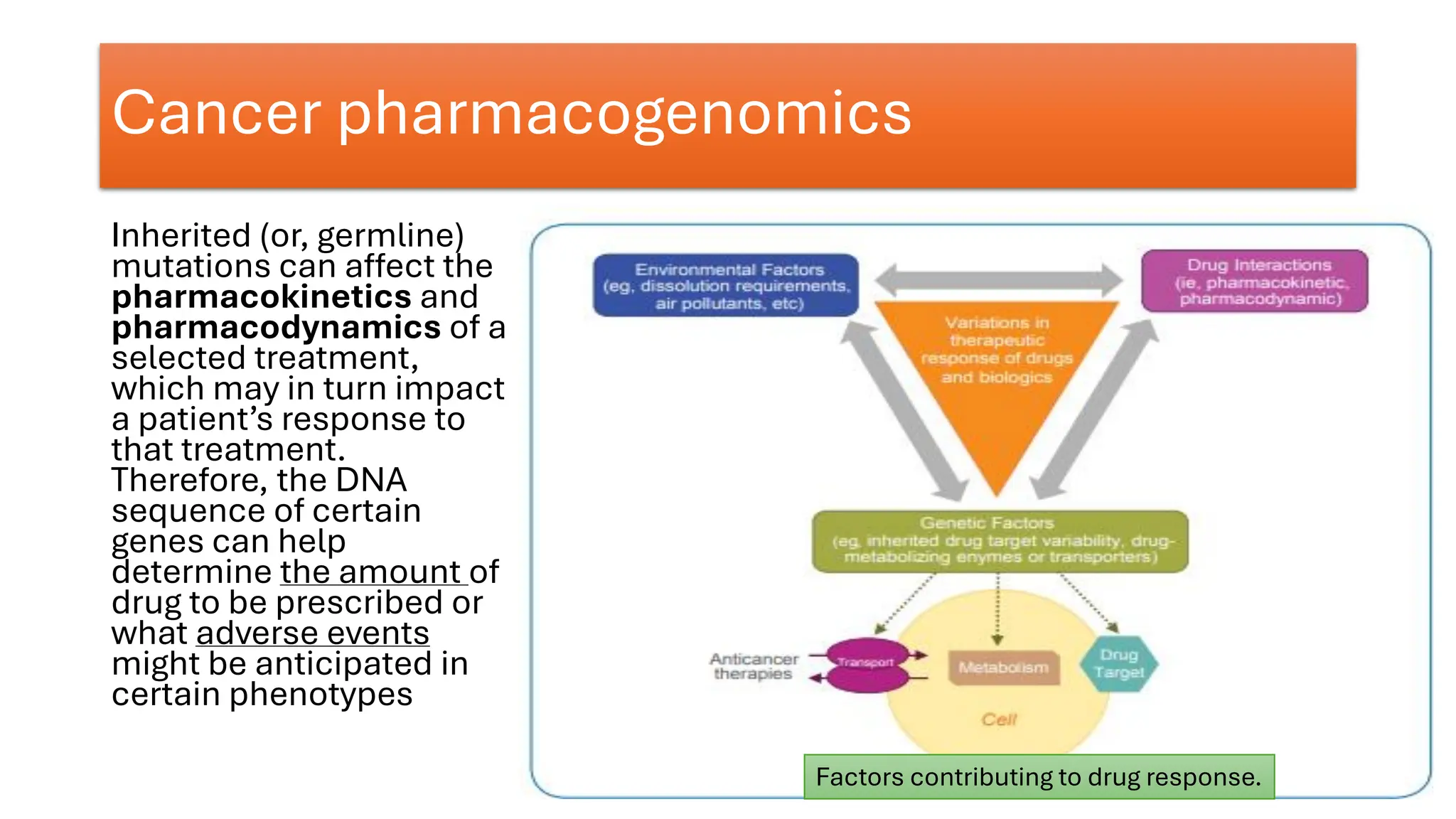
The document outlines the use of Next Generation Sequencing (NGS) at the KCCC for personalized medicine in oncology, detailing its workflow, clinical applications, and the advantages over traditional sequencing methods. NGS enables faster analysis of multiple genes and mutations, supports precision oncology, and has case studies demonstrating its effectiveness in improving patient outcomes. Furthermore, it highlights the challenges of interpreting data and the role of liquid biopsies in cancer diagnostics.