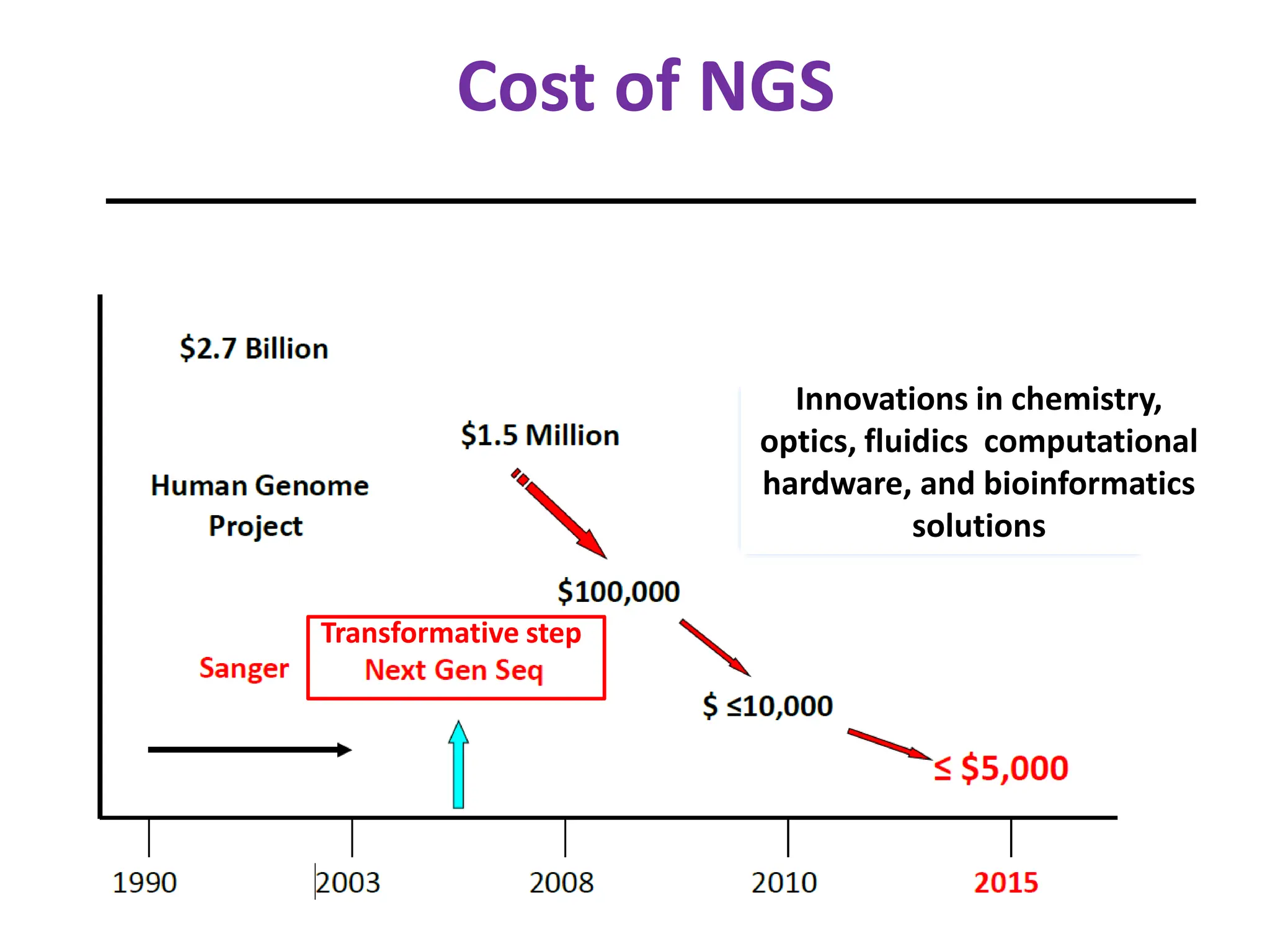
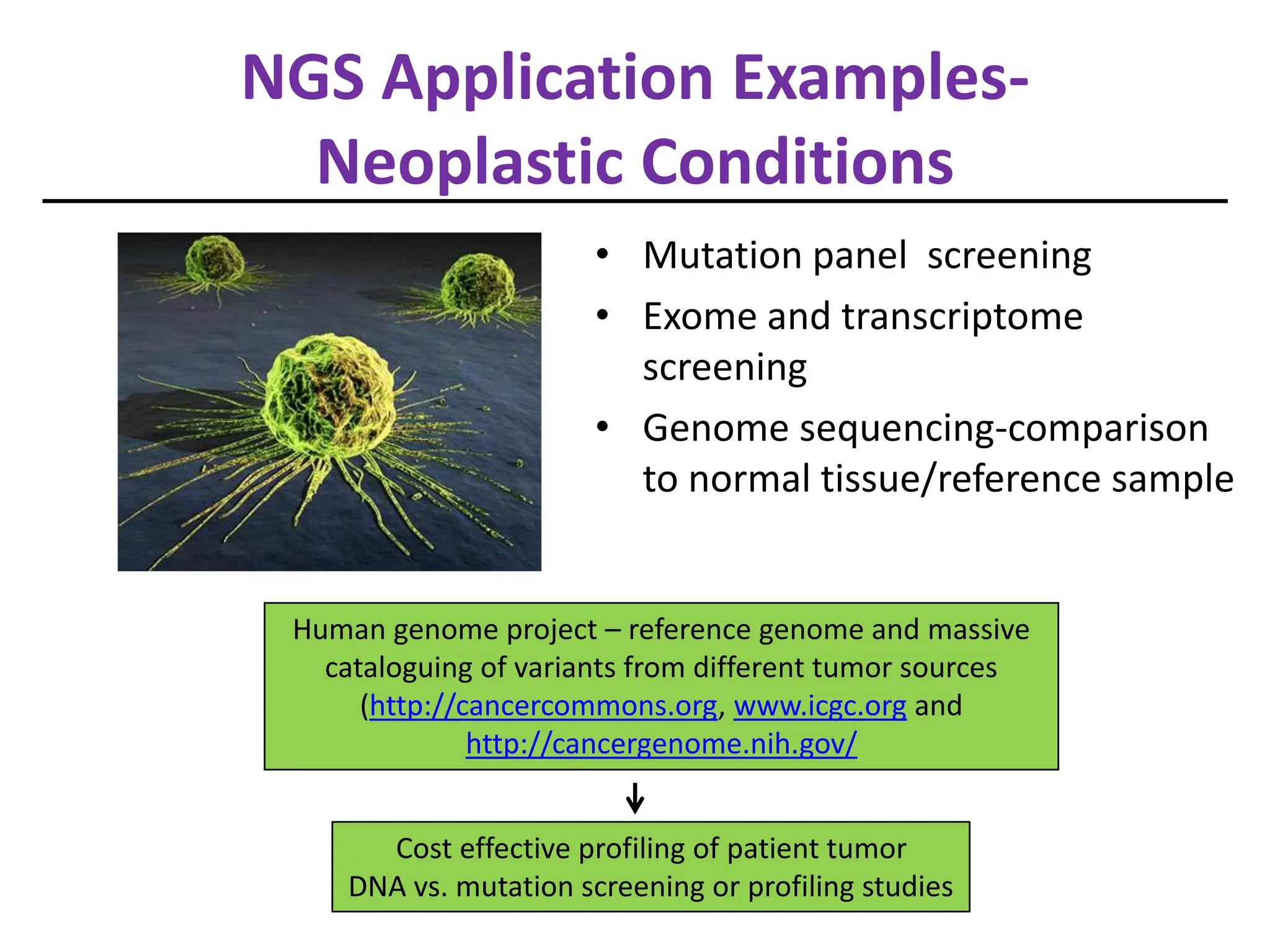
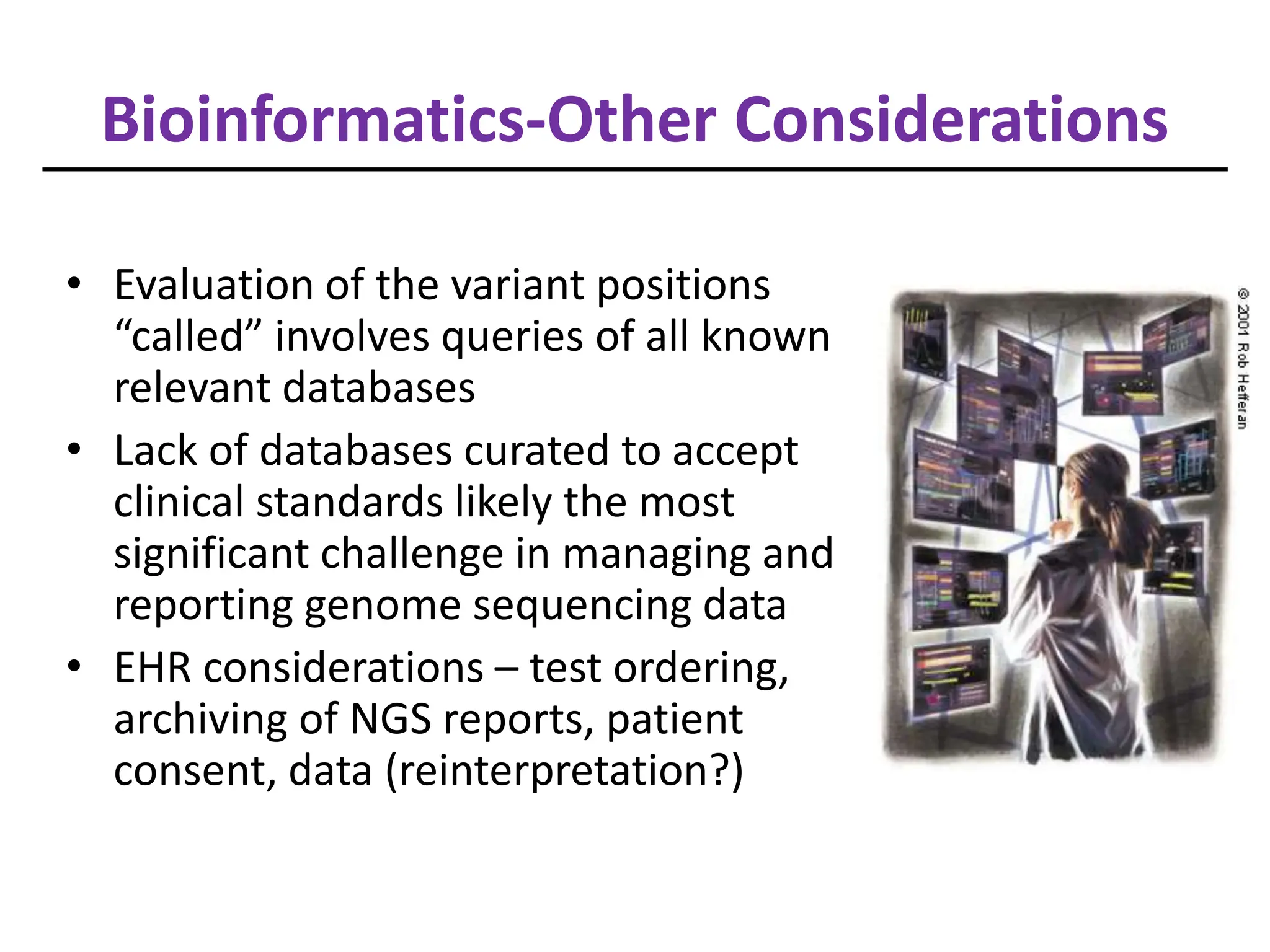
NGS has arrived in the clinic and provides opportunities for improved diagnosis and treatment of inherited conditions and cancers. However, there are also challenges to address regarding clinical validity, informed consent, analytical validation, regulation, data analysis, interpretation and education. Expert teams are needed to maximize clinical utility while minimizing risks like incidental findings and uncertainty. Guidelines are being developed to help optimize use of this transformative technology.