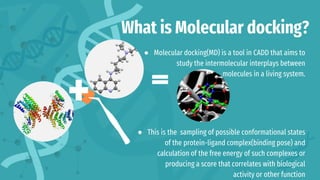
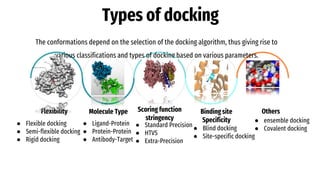
The document provides an introduction to molecular docking (MD), a computational tool used in drug discovery to study the interactions between molecules. It outlines the theory, applications, and limitations of MD, emphasizing its role in biomedical research and the challenges faced, such as inaccuracy in energy calculations and limited sampling. The document also highlights the historical development of MD and its significance in understanding biological processes.