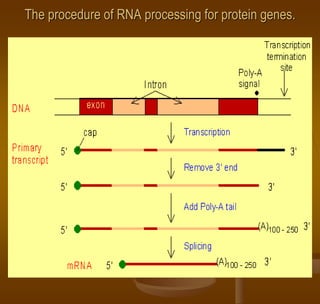
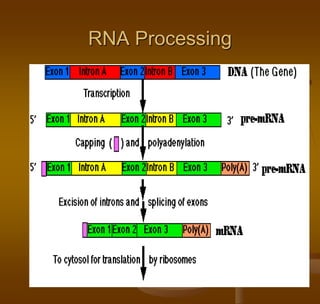
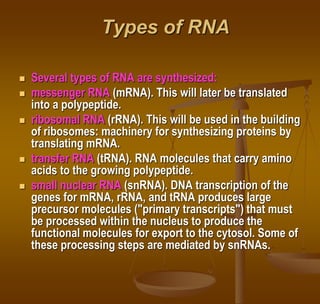
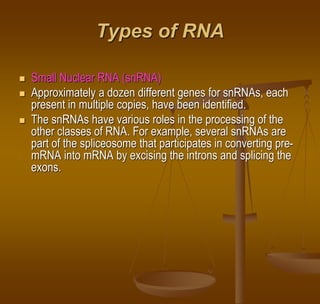
The document discusses RNA processing, which involves several key steps:
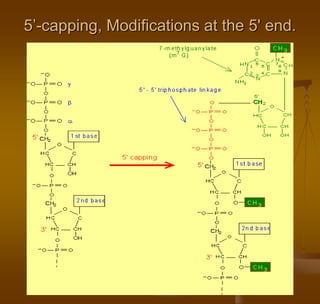
1) Capping at the 5' end, which protects RNA from degradation and aids in transport and splicing.
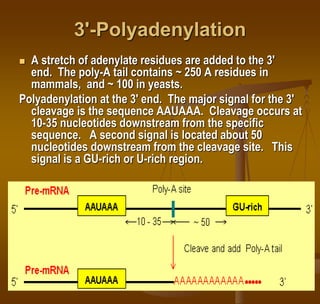
2) Polyadenylation at the 3' end, which also protects RNA and aids in transport.
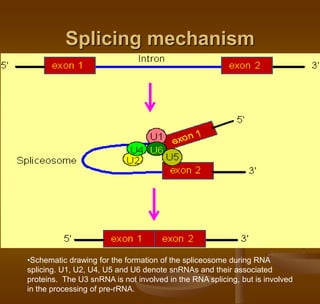
3) Splicing, which removes introns and joins exons using a spliceosome complex.
The end result is mature mRNA that can be translated into protein.