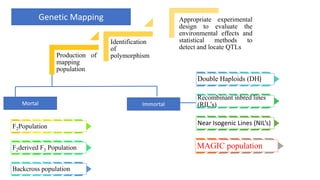
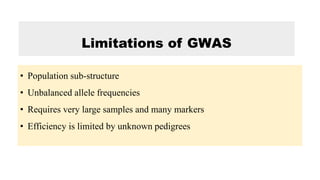
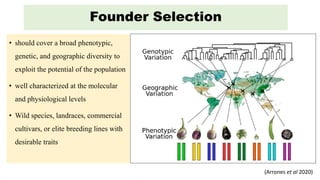
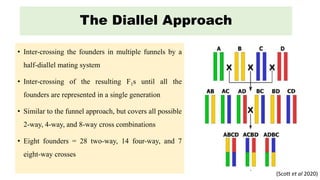
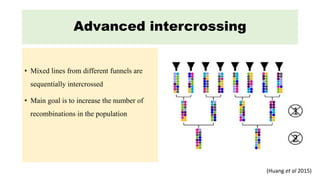
The document describes the use of MAGIC (Multi-parent Advanced Generation Inter-Cross) populations for vegetable improvement and QTL mapping. MAGIC populations have increased genetic diversity compared to biparental populations due to the use of multiple founders. They allow for improved QTL mapping resolution and detection. Case studies in tomato, brinjal, and common bean are described where MAGIC populations were developed and used to map QTL for traits like anthocyanin content and drought tolerance. Founder lines are intercrossed in funnel or diallel designs over multiple generations to generate immortal RIL populations with equal proportions of founder genomes.