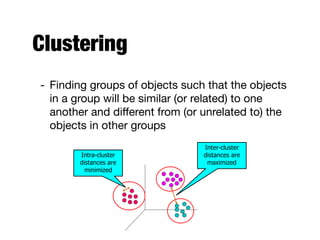
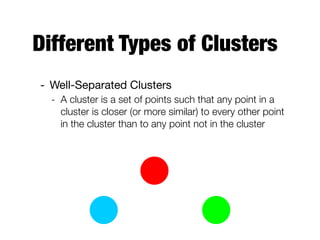
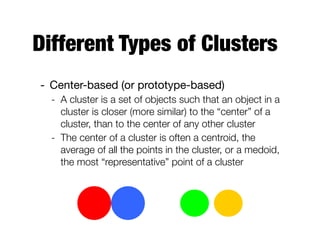
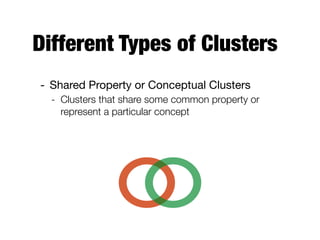
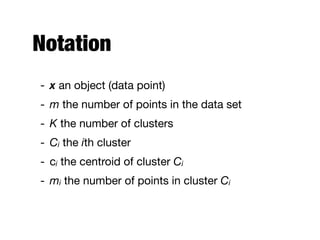
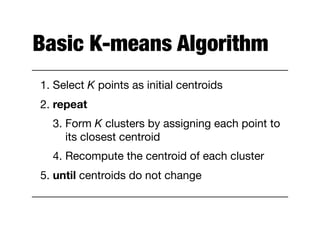
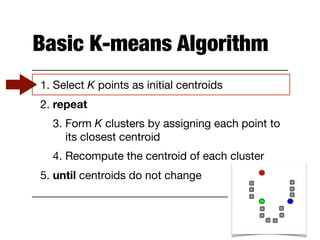
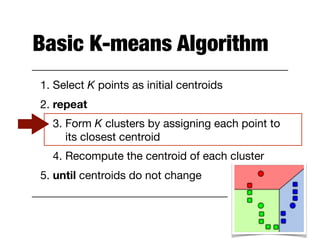
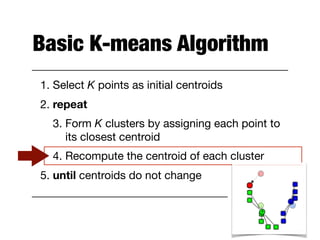
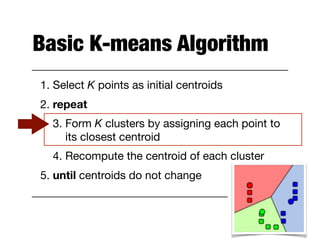
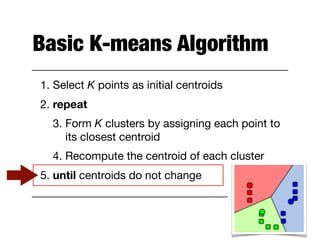
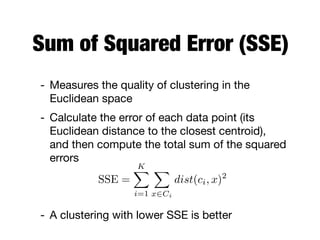
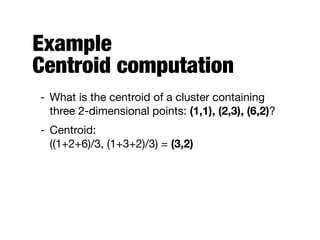
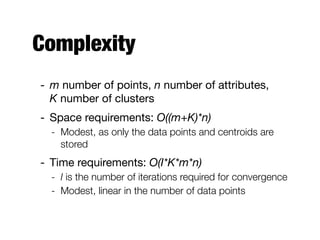
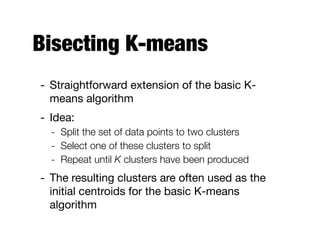
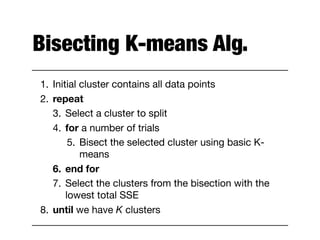
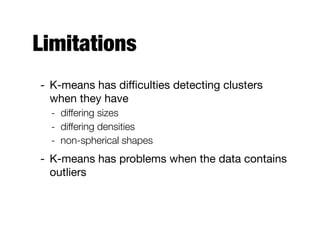
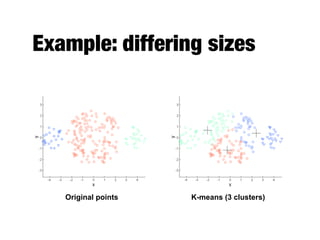
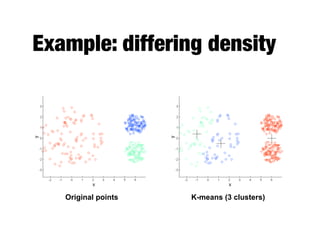
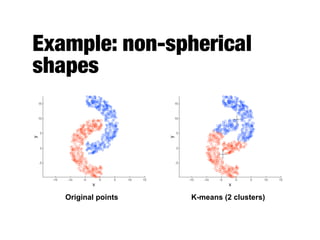
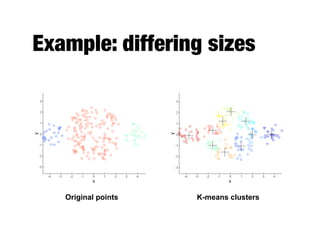
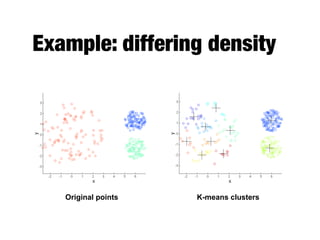
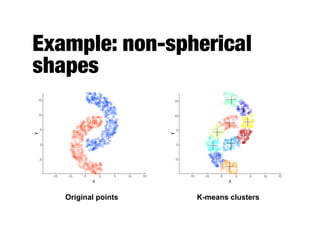
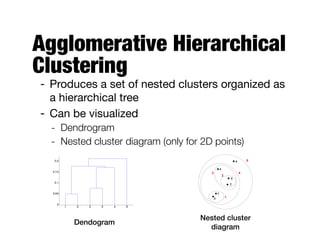
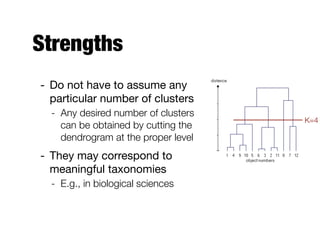
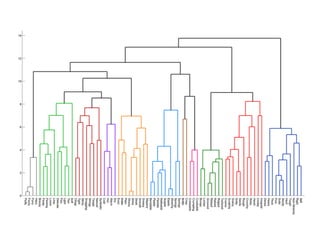
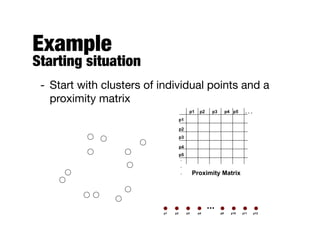
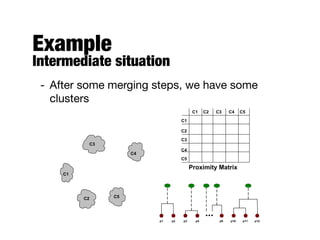
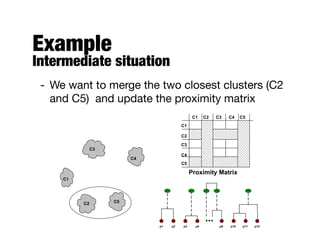
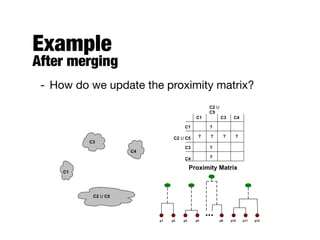
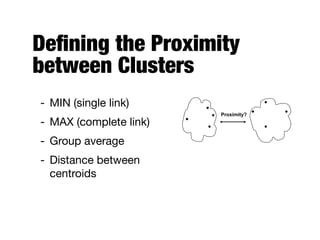
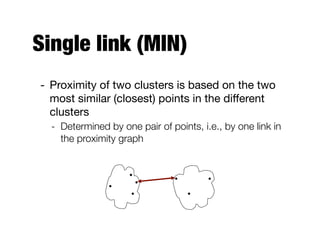
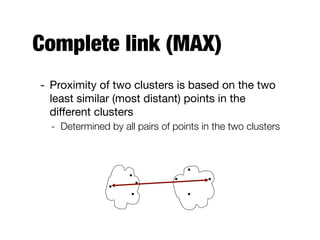
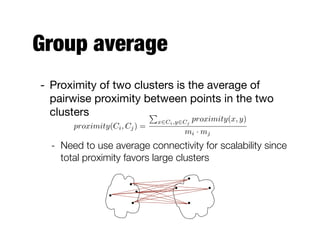
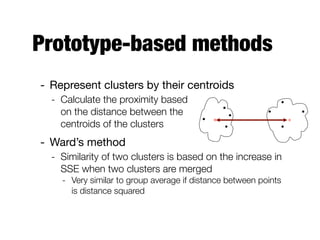
Hierarchical clustering builds clusters hierarchically, by either merging or splitting clusters at each step. Agglomerative hierarchical clustering starts with each point as a separate cluster and successively merges the closest clusters based on a defined proximity measure between clusters. This results in a dendrogram showing the nested clustering structure. The basic algorithm computes a proximity matrix, then repeatedly merges the closest pair of clusters and updates the matrix until all points are in one cluster.