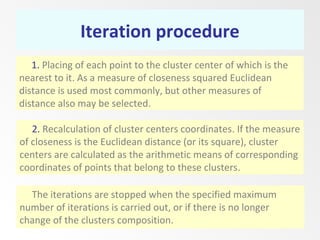
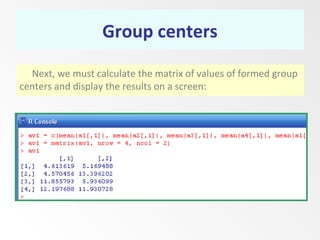
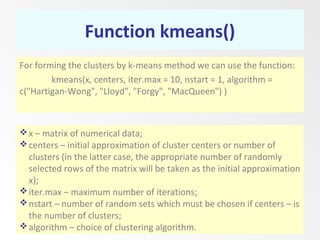
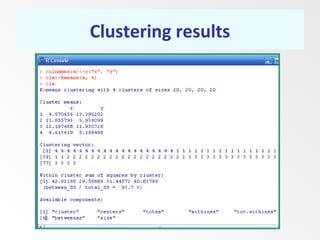
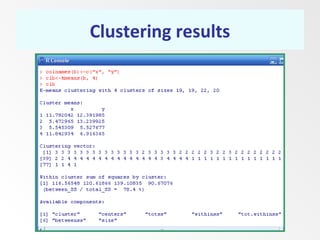
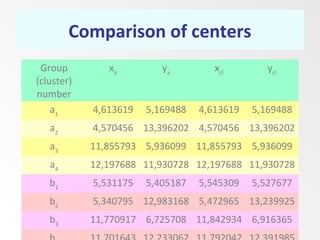
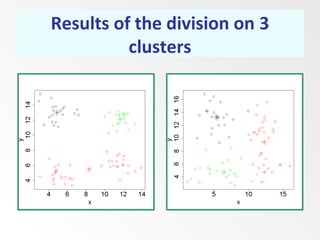
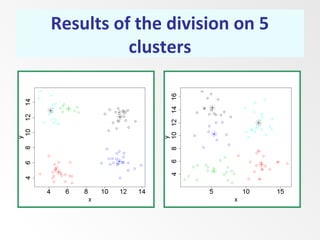
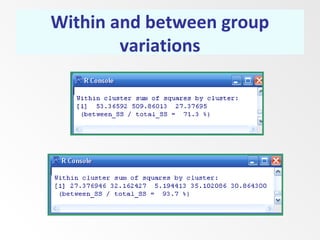
This document discusses the k-means clustering method. It begins by defining the problem of cluster analysis as dividing data points into groups to minimize the sum of distances between points and their assigned cluster centers. It then describes the main k-means algorithms and outlines the iterative process of assigning points to the nearest cluster center and recalculating the centers. Finally, it provides an example applying k-means clustering to sample data and analyzing the results.