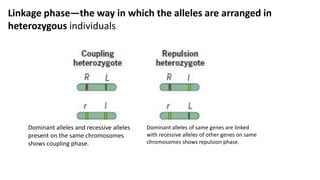
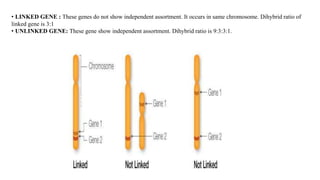
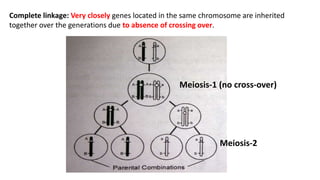
1) Linkage is the tendency of genes located near each other on the same chromosome to be inherited together during meiosis. Genes farther apart on a chromosome assort independently according to Mendel's law of independent assortment.
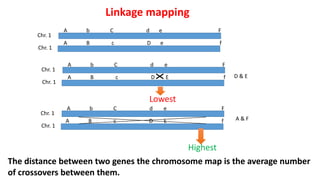
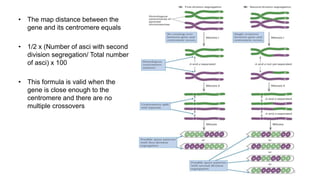
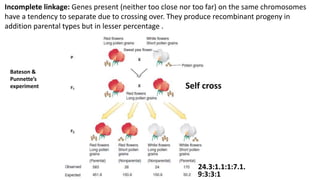
2) The strength of linkage depends on the physical distance between genes - closely located genes show complete linkage while genes farther apart show incomplete linkage with crossover occurring.
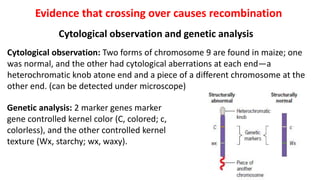
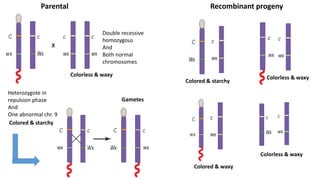
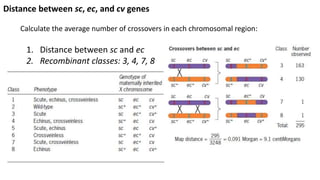
3) Tetrad analysis in fungi allows the direct observation of linkage and estimation of recombination frequencies between genes and centromeres.



![Frequency (percentage) of recombination (RF) ranges from 0 to
0.5 (0-50%)
1. The more RF is close to 50% the more chances is there for the genes to be
unlinked (independent assortment).
Mendel’s experiment RF= 9:3:3:1 [6/16 x100 = 38%]
2. The more RF is close to 0% the more chances is there for the genes to be
tightly linked (medium or close linkage).
Bateson & Punnette’s experiment RF = [0.08 OR 8%]](https://image.slidesharecdn.com/linkagecrossingover-221127210805-dfa82bee/85/Linkage_Crossing_over-pptx-8-320.jpg)