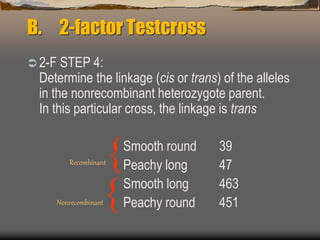
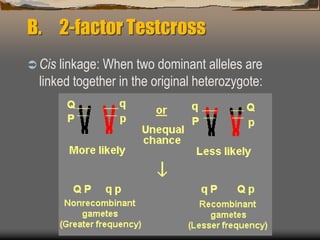
The document discusses linkage mapping in genetics, explaining the physical basis, and methodologies for 2-factor and 3-factor testcrosses. It describes how recombination frequencies relate to gene distances and details the steps to calculate these distances using both types of testcrosses, including an example with tomato plants and maize. Additionally, it covers concepts of interference and the coefficient of coincidence in relation to double crossovers.