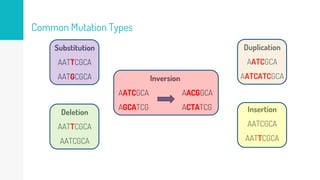
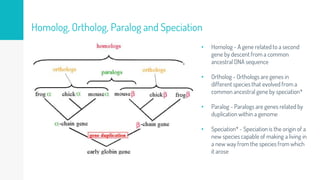
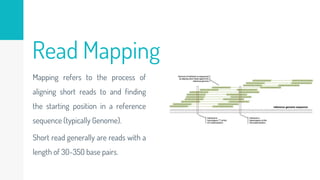
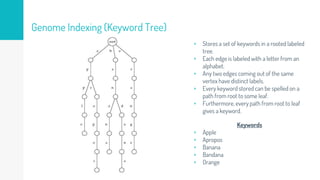
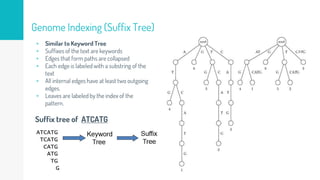
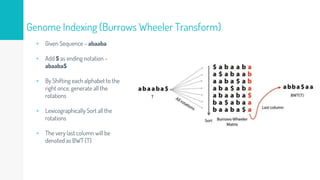
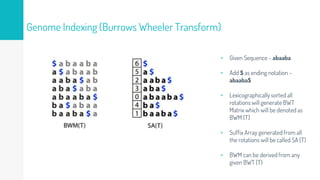
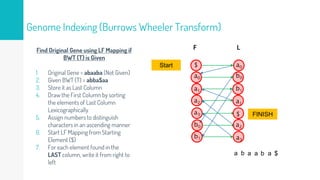
Gene duplication refers to the duplication of a region of DNA containing a gene. This is a major mechanism of generating new genetic material during evolution. Read mapping involves aligning short DNA reads to a reference genome sequence to determine their positions. Several data structures like suffix trees, suffix arrays, and the Burrows-Wheeler transform are used to index genomes for efficient read mapping.