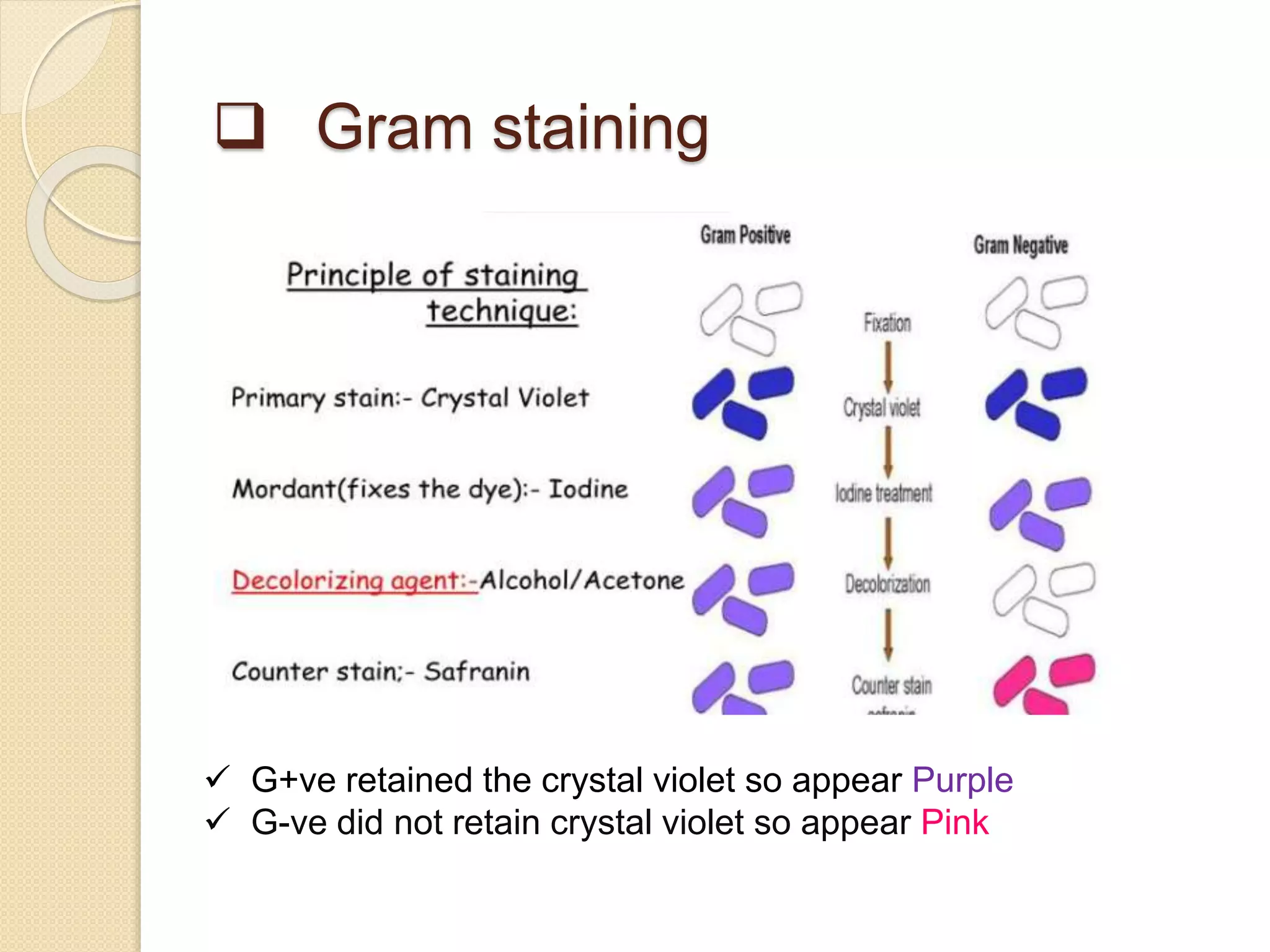
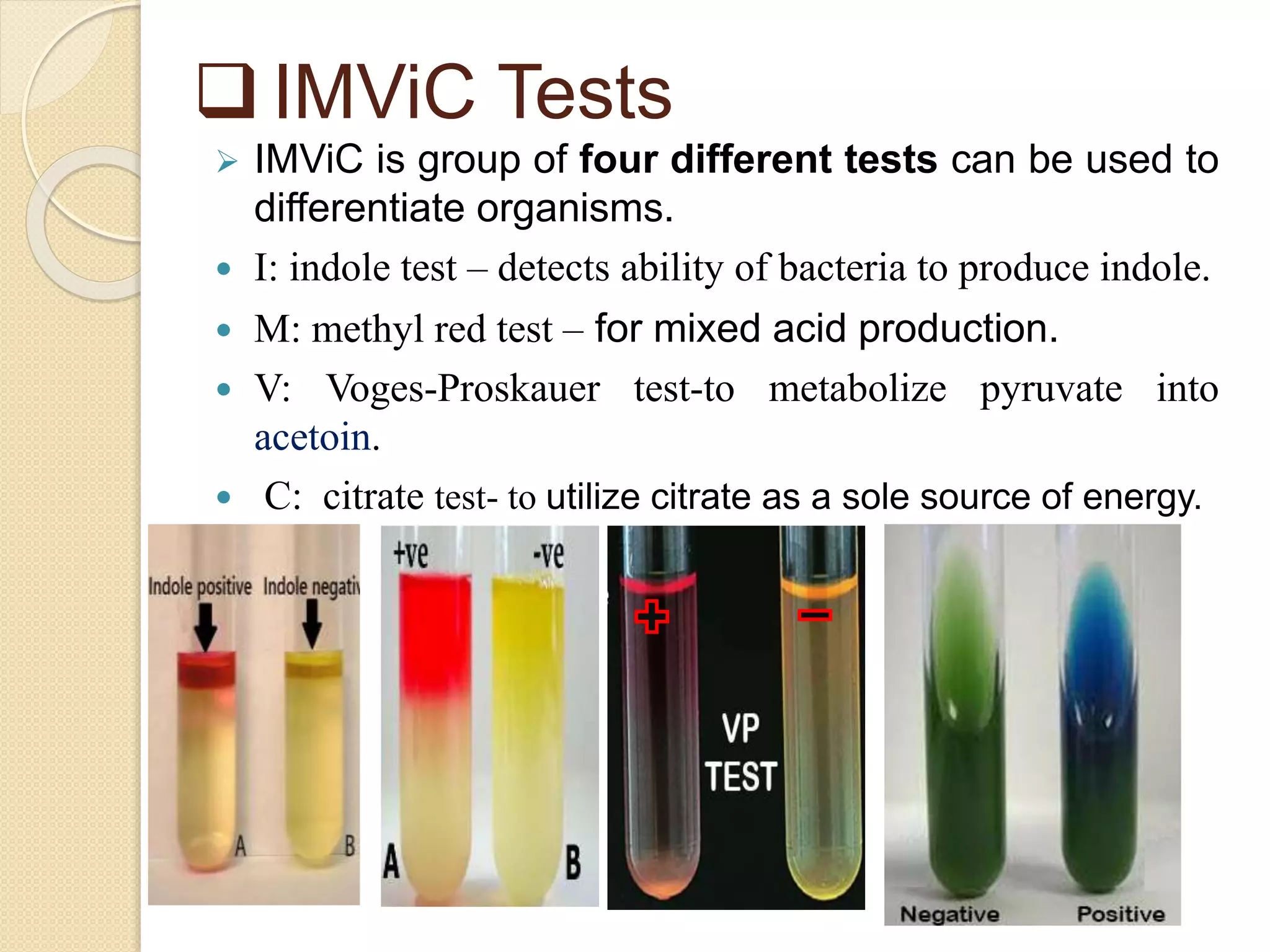
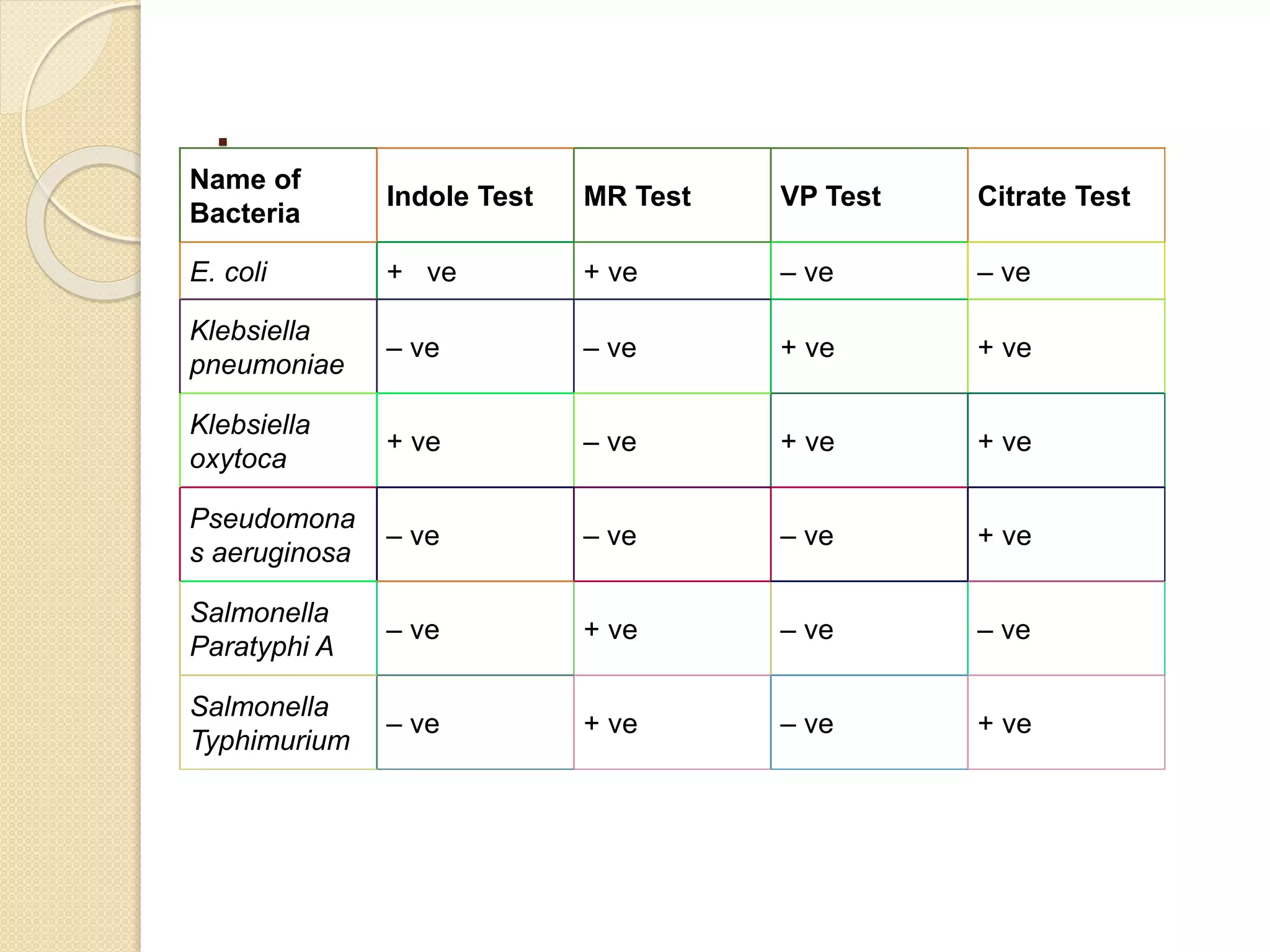
The document discusses techniques for classification and identification of microorganisms, focusing on methods such as morphological, physiological, biochemical, immunological, and molecular approaches. It details various staining techniques, including simple and differential staining, and specific biochemical tests that help identify bacterial species based on metabolic needs. Additionally, it covers molecular methods like ribotyping and PCR for microbial DNA evaluation and provides references for further reading.