1. Genetics plays an important role in medicine through studies of inheritance patterns, gene mapping, analysis of disease mechanisms, and diagnosis/treatment of genetic diseases like gene therapy.
2. DNA isolation involves extracting DNA from samples and separating it from other cell components. It is used for scientific research, medicine like outbreak tracing, and forensic science like identification. Various methods disrupt cells, remove proteins, and recover DNA.
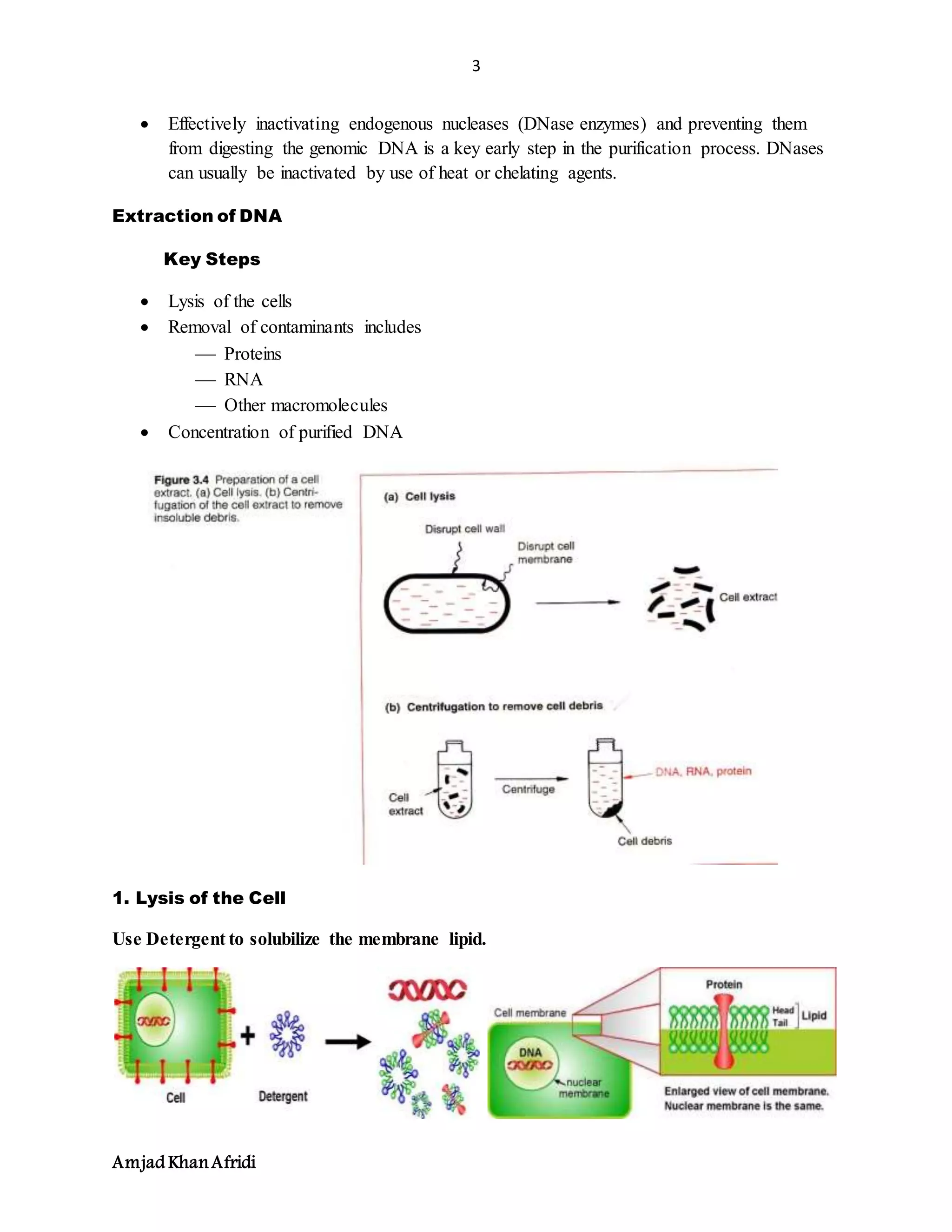
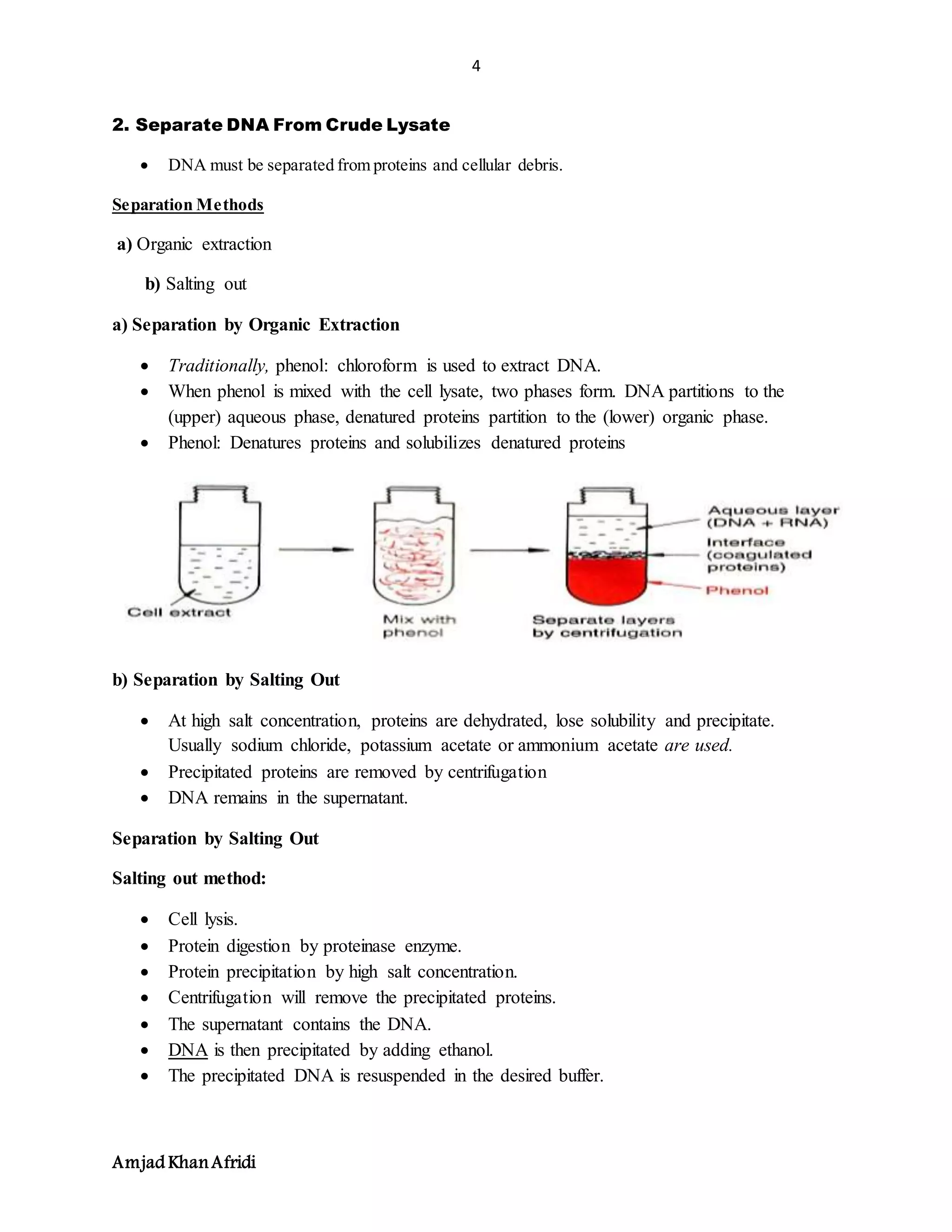
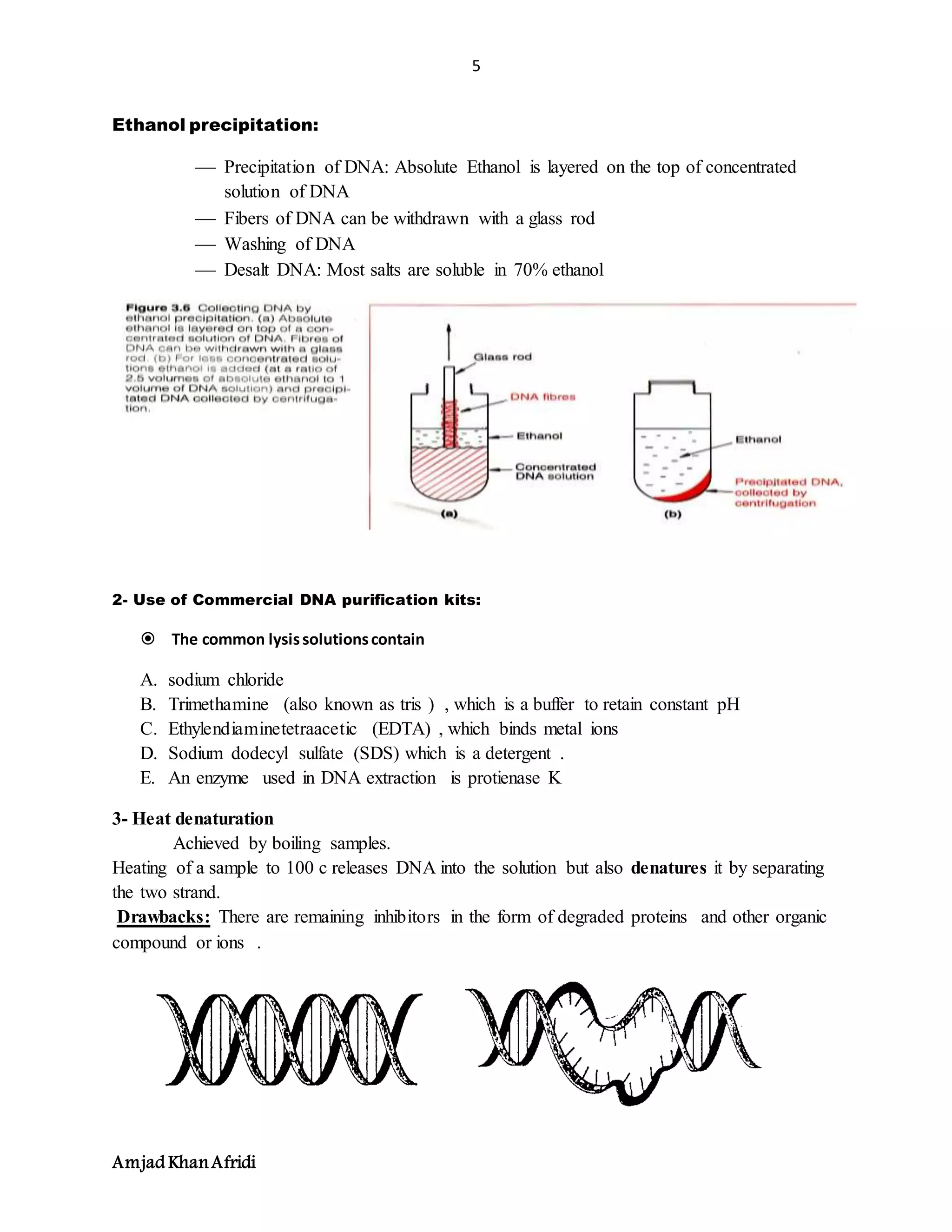
3. DNA purification separates DNA from other cell components and avoids fragmentation. Key steps include cell lysis, removal of contaminants like proteins and RNA, and concentrating the purified DNA.