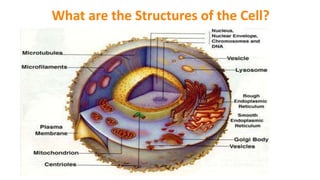
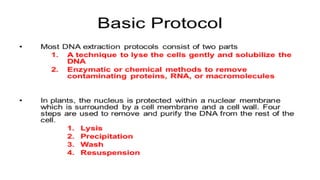
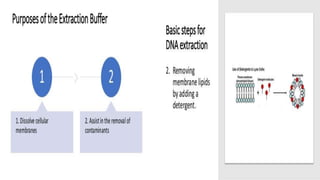
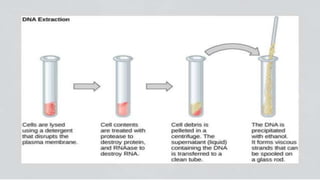
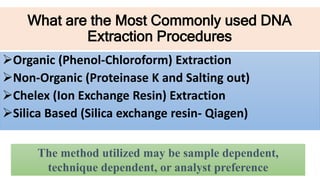
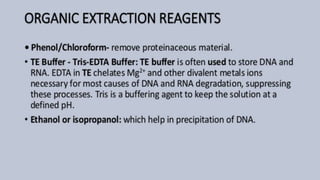
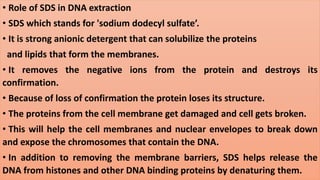
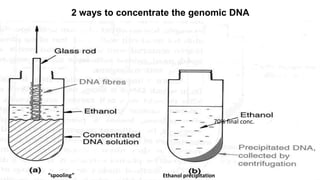
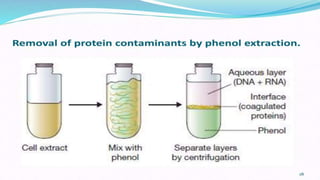
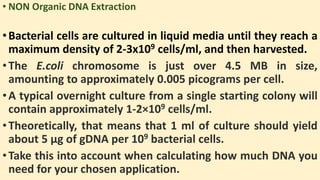
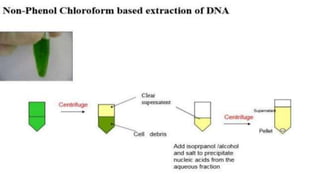
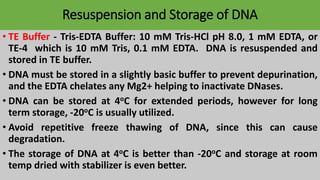
The document discusses various methods for extracting DNA from cells and tissues. It describes key steps in DNA extraction procedures including cell lysis to release DNA, removal of contaminants, DNA precipitation and purification. Common techniques discussed are organic extraction using phenol-chloroform, non-organic extraction using proteinase K and salting out, and silica-based extraction. The document also covers concentrating extracted DNA using alcohol precipitation and resuspension of purified DNA in TE buffer for storage.