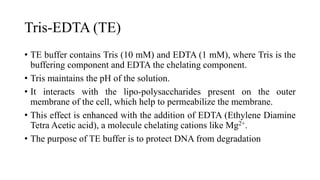
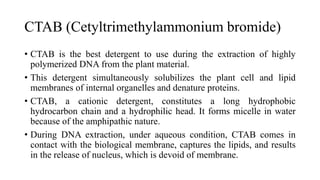
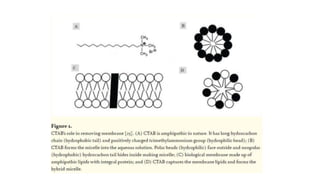
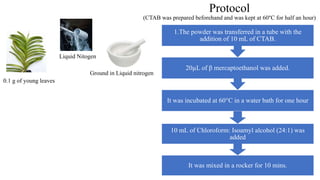
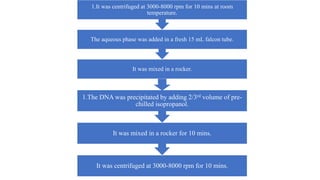
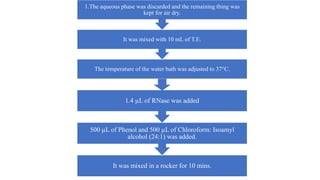
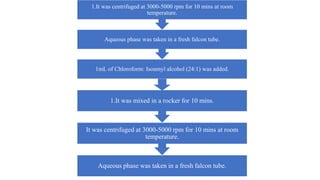
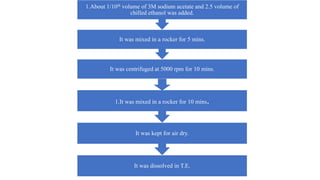
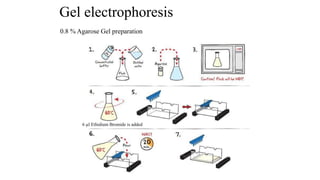
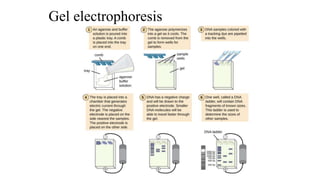
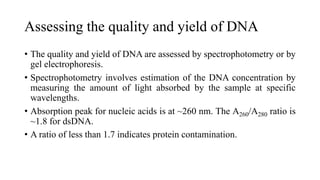
DNA extraction is a method to purify DNA from a sample by separating it from other cellular components using physical and chemical methods. It involves disrupting cell walls and membranes to release DNA, then precipitating the DNA and removing contaminants like proteins, lipids, and RNA. Common extraction methods use chemicals like phenol, chloroform, and isopropanol to separate DNA, which is then analyzed on a gel for quality and yield assessment or used in applications like forensics, ancestry tracking, medical testing, and genetic engineering.