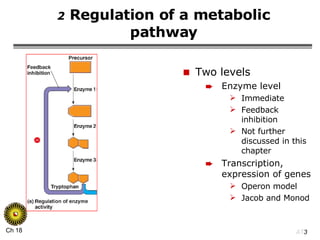
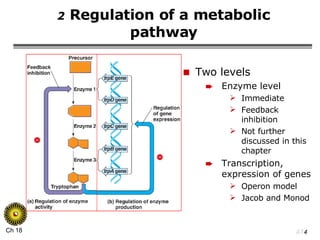
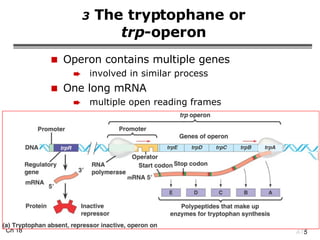
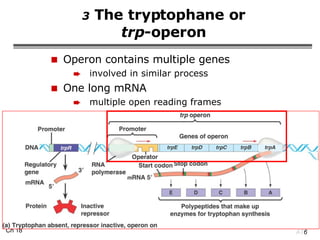
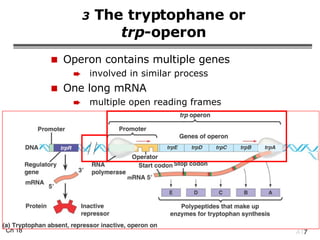
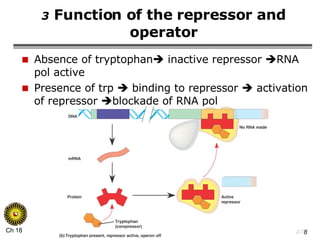
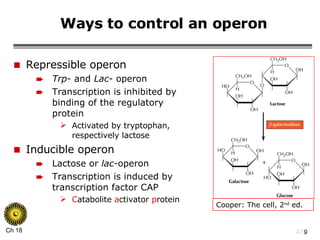
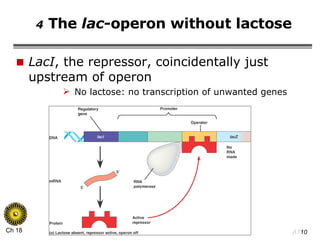
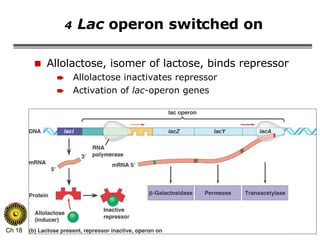
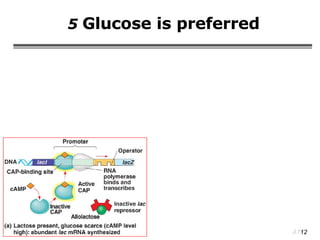
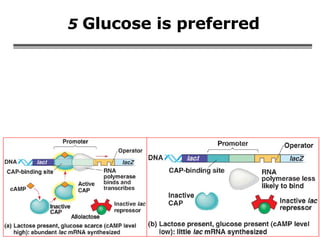
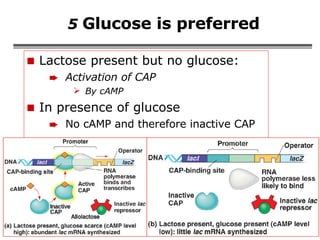
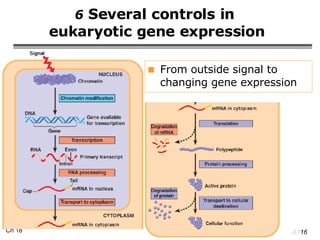
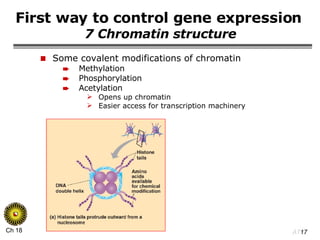
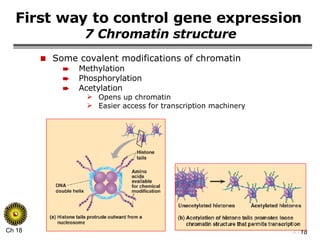
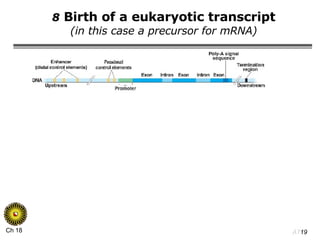
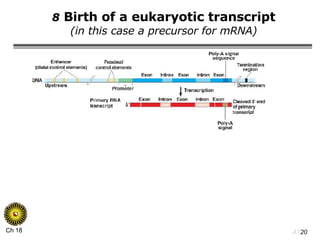
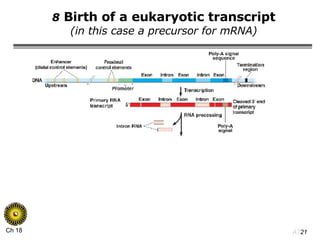
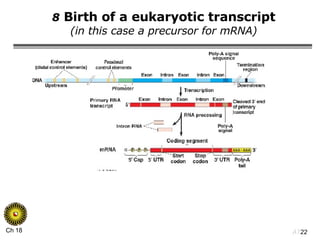
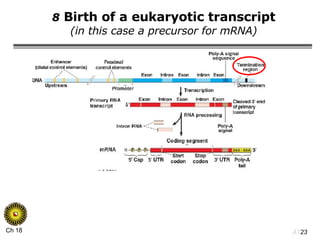
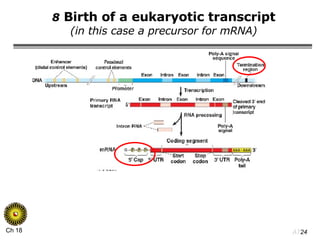
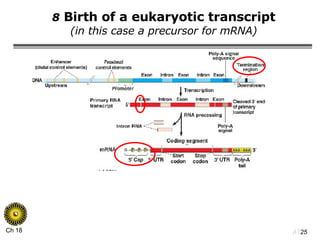
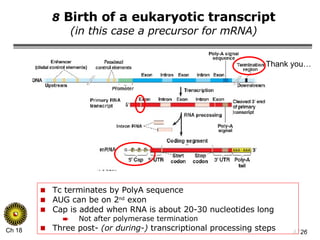
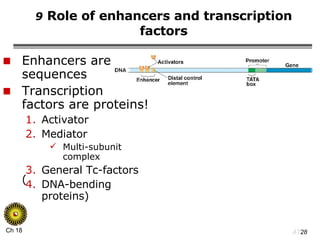
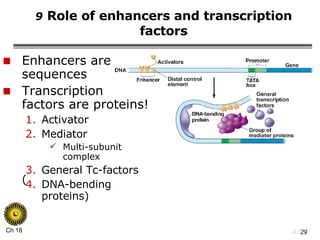
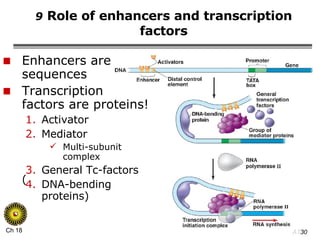
The document summarizes regulation of gene expression in bacteria and eukaryotes. It discusses the trp and lac operons in bacteria, which regulate gene expression in response to tryptophan and lactose levels. In eukaryotes, gene expression is controlled by chromatin structure, transcription factors that bind enhancers and recruit RNA polymerase, and post-transcriptional processing of transcripts.