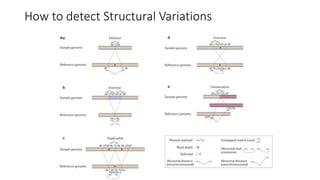
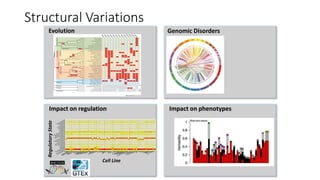
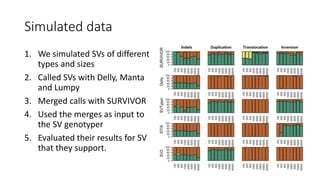
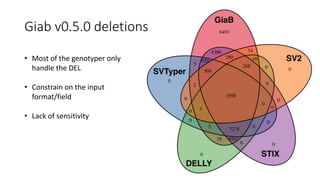
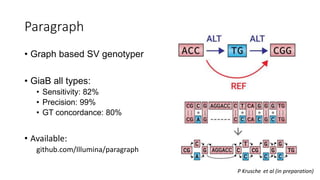
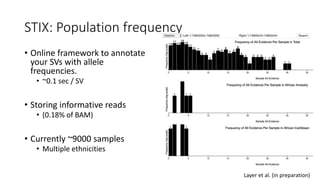
The document discusses challenges in computational genotyping of structural variations for clinical diagnosis. It describes assessing accuracy of structural variation calls, challenges in functional interpretation, and how to call structural variations in routine scans. It also discusses using simulated and benchmark data to evaluate structural variation genotypers and their ability to sensitively and precisely genotype deletions. A new graph-based genotyper called Paragraph is highlighted for its ability to genotype multiple structural variation types with high accuracy based on benchmarking.